Introduction #
The authors of the DeepBacs: Bacterial Image Analysis Using Open-source Deep Learning Approaches biomedical project highlight the rapid transformation of microscopy through Deep Learning, which has enabled efficient analysis of complex data and often surpassed the performance of classical algorithms. As a result, there has been a significant effort to develop user-friendly tools that allow biomedical researchers, with limited computer science backgrounds, to leverage DL effectively. While such approaches have primarily focused on analyzing microscopy images from eukaryotic samples, they have been underutilized in microbiology.
To support researchers in their training process, the authors provide a specifically designed database of training and testing data, allowing bacteriologists to quickly explore how to analyze their data using DL techniques. The ultimate goal is to foster the efficient application of DL in microbiology, leading to the creation of novel tools for bacterial cell biology and antibiotic research.
The presented dataset contains training and test images of E. coli cells treated with different antibiotics for antibiotic phenotyping:
| Characteristic | Value |
|---|---|
| Data type | Paired microscopy images (confocal fluorescence) and manual annotations |
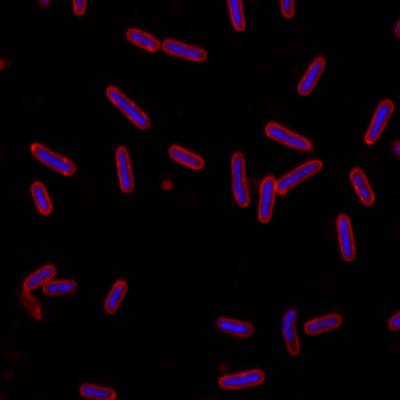
| Microscopy data type | Confocal fluorescence images of fixed E. coli cells stained for membrane (Nile Red) and DNA (DAPI) paired with annotations in PASCAL VOC format |
| Microscope | Zeiss LSM710 confocal microscope with a Plan-Apo 63x oil objective (1.4 NA) |
| Cell type | Chemically fixed E. coli NO34 cells (MreB-sfGFPsw, kindly provided by Zemer Gitai) (untreated or drug-treated); |
| Image size | 400 x 400 px² (Pixel size: 84 nm) |
Summary #
DeepBacs: Escherichia Coli Antibiotic Phenotyping Object Detection Dataset and YOLOv2 Model is a dataset for an object detection task. It is used in the biomedical research.
The dataset consists of 235 images with 3564 labeled objects belonging to 8 different classes including Oblique, Vesicle, Nalidixate, and other: Rifampicin, CAM, Control, Mecillinam, and MP265.
Images in the DeepBacs E. Coli dataset have bounding box annotations. All images are labeled (i.e. with annotations). There are 3 splits in the dataset: train (153 images), test_individual_treatment (50 images), and test_stitched_images (32 images). The dataset was released in 2021 by the Institute of Physical and Theoretical Chemistry, Goethe-University Frankfurt, Germany.
Here is the visualized example grid with annotations:
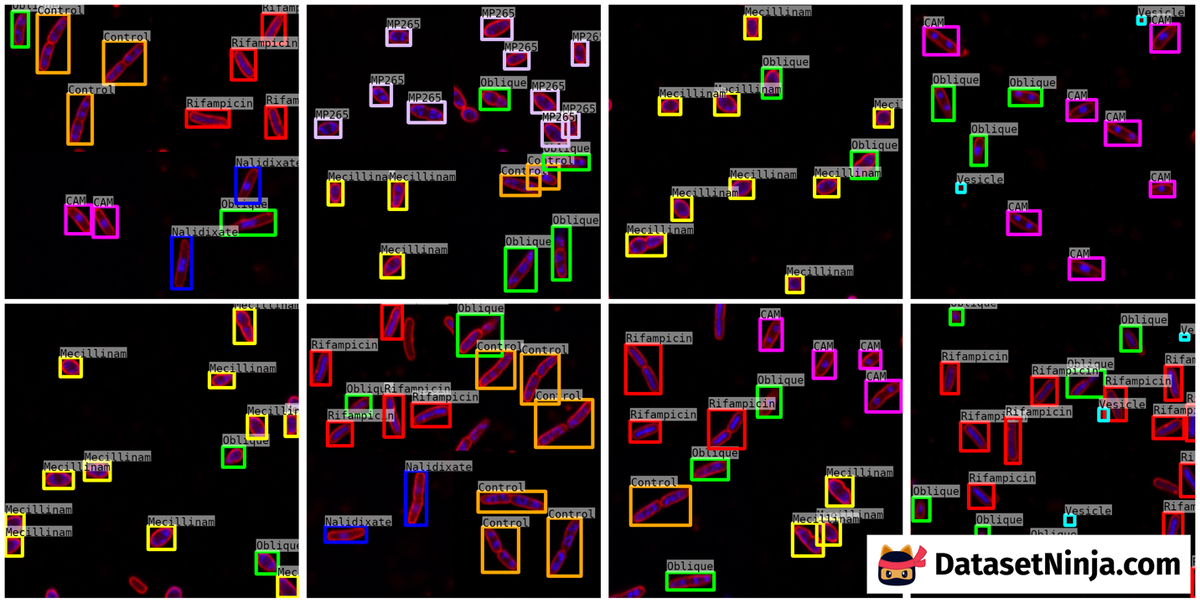
Explore #
DeepBacs E. Coli dataset has 235 images. Click on one of the examples below or open "Explore" tool anytime you need to view dataset images with annotations. This tool has extended visualization capabilities like zoom, translation, objects table, custom filters and more. Hover the mouse over the images to hide or show annotations.




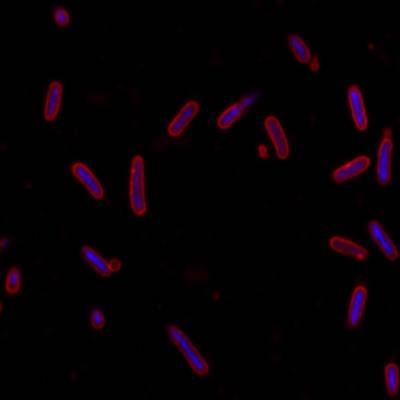

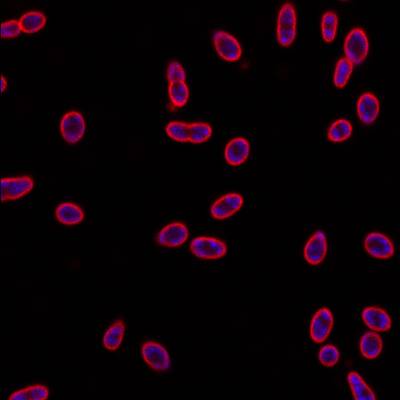

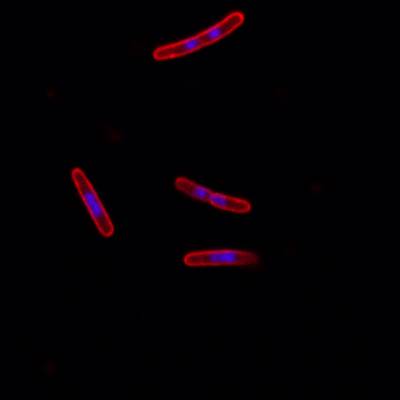

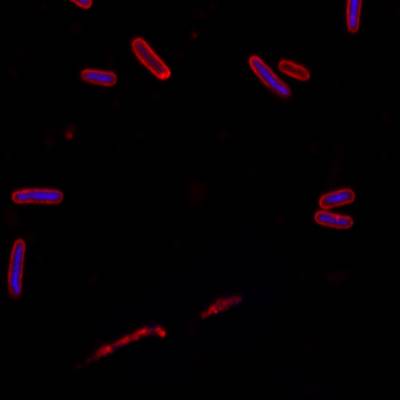

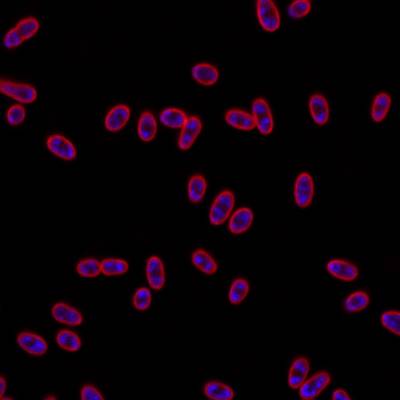

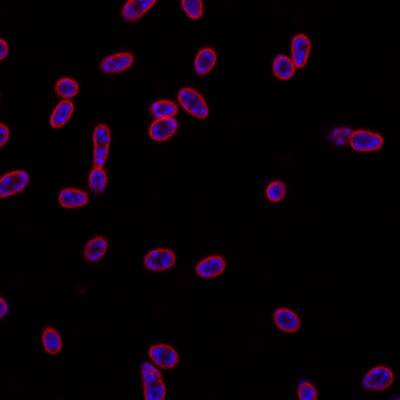



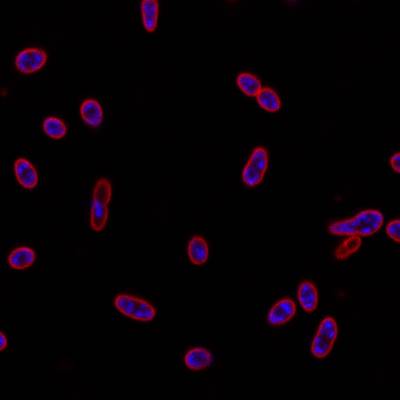

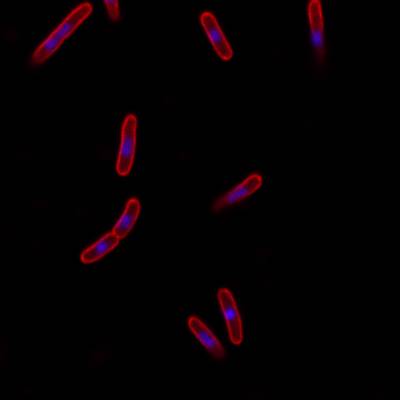

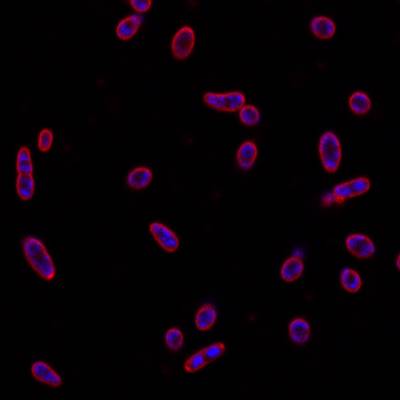

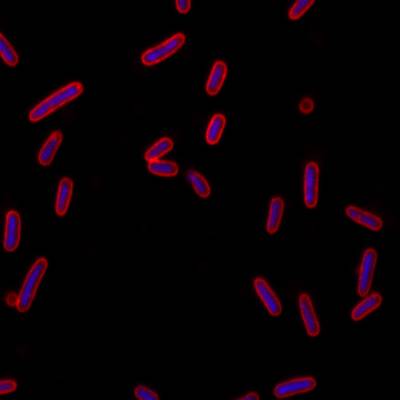

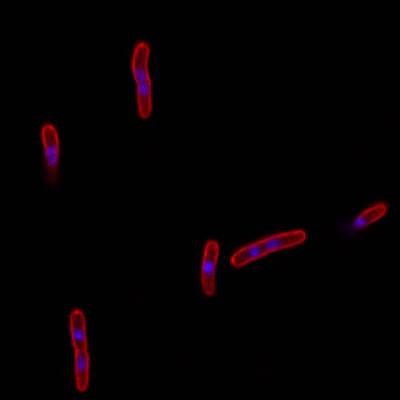

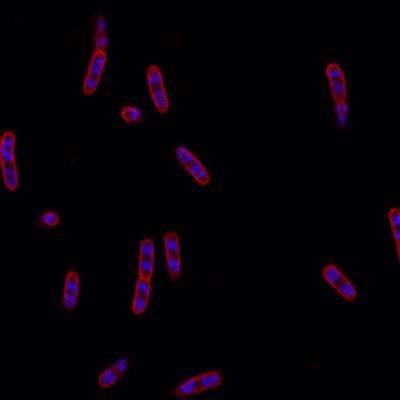



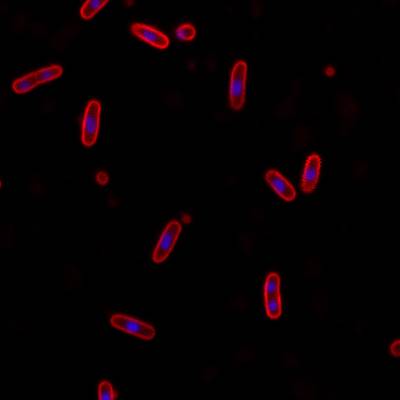

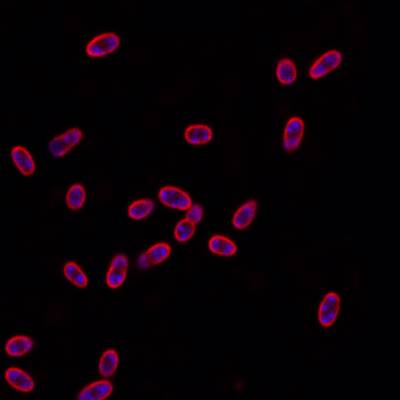

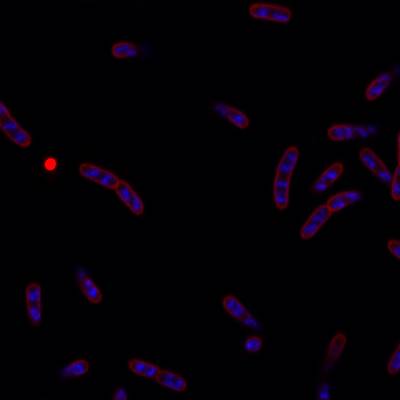

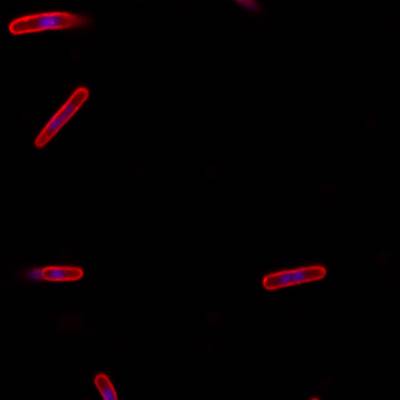

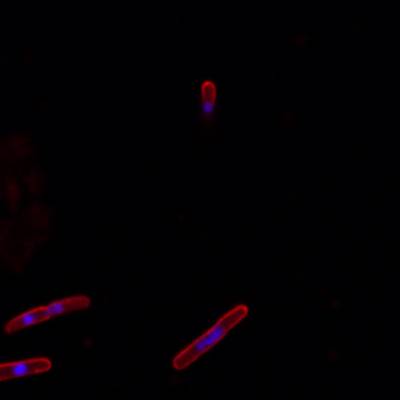

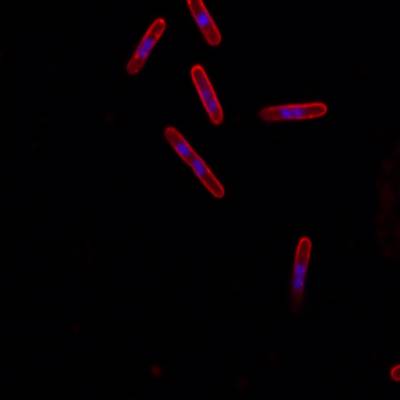

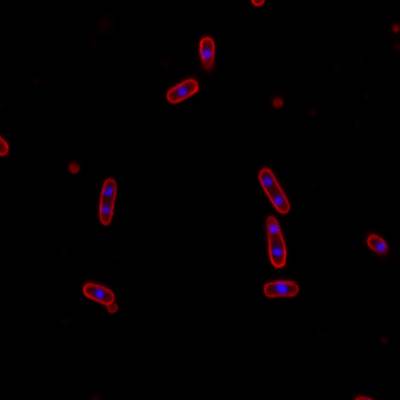

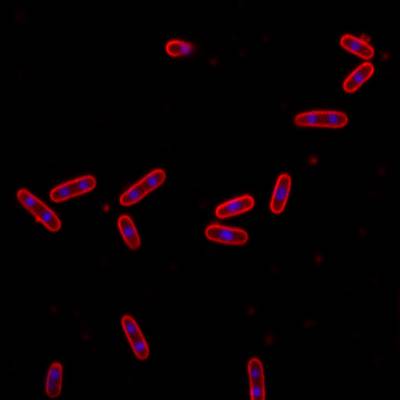

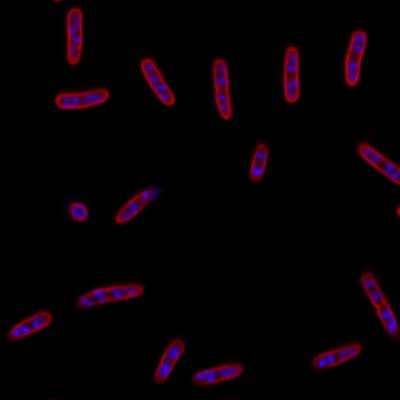

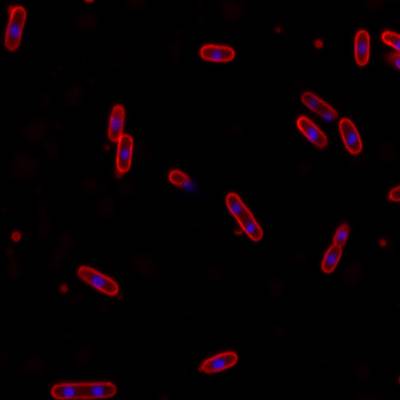

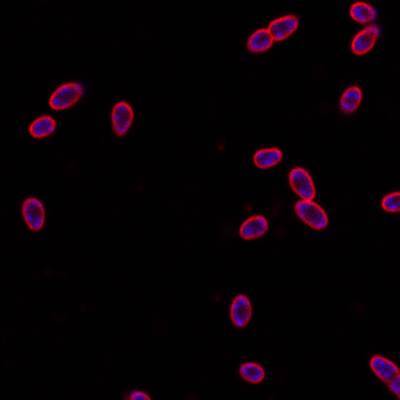

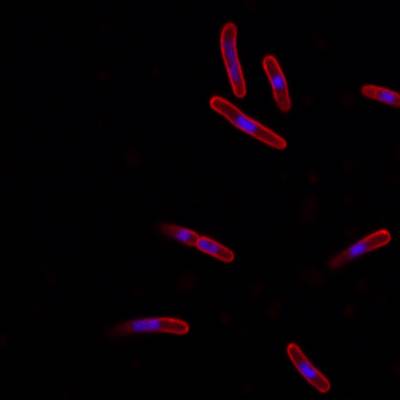

Class balance #
There are 8 annotation classes in the dataset. Find the general statistics and balances for every class in the table below. Click any row to preview images that have labels of the selected class. Sort by column to find the most rare or prevalent classes.
Class ㅤ | Images ㅤ | Objects ㅤ | Count on image average | Area on image average |
|---|---|---|---|---|
Oblique➔ rectangle | 201 | 789 | 3.93 | 2.98% |
Vesicle➔ rectangle | 100 | 393 | 3.93 | 0.45% |
Nalidixate➔ rectangle | 64 | 209 | 3.27 | 4.64% |
Rifampicin➔ rectangle | 56 | 597 | 10.66 | 8.82% |
CAM➔ rectangle | 52 | 374 | 7.19 | 5.82% |
Control➔ rectangle | 48 | 308 | 6.42 | 7.88% |
Mecillinam➔ rectangle | 47 | 484 | 10.3 | 6.73% |
MP265➔ rectangle | 37 | 410 | 11.08 | 6.5% |
Co-occurrence matrix #
Co-occurrence matrix is an extremely valuable tool that shows you the images for every pair of classes: how many images have objects of both classes at the same time. If you click any cell, you will see those images. We added the tooltip with an explanation for every cell for your convenience, just hover the mouse over a cell to preview the description.
Images #
Explore every single image in the dataset with respect to the number of annotations of each class it has. Click a row to preview selected image. Sort by any column to find anomalies and edge cases. Use horizontal scroll if the table has many columns for a large number of classes in the dataset.
Object distribution #
Interactive heatmap chart for every class with object distribution shows how many images are in the dataset with a certain number of objects of a specific class. Users can click cell and see the list of all corresponding images.
Class sizes #
The table below gives various size properties of objects for every class. Click a row to see the image with annotations of the selected class. Sort columns to find classes with the smallest or largest objects or understand the size differences between classes.
Class | Object count | Avg area | Max area | Min area | Min height | Min height | Max height | Max height | Avg height | Avg height | Min width | Min width | Max width | Max width |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Oblique rectangle | 789 | 0.76% | 4.84% | 0.05% | 10px | 2.5% | 106px | 26.5% | 35px | 8.66% | 8px | 2% | 97px | 24.25% |
Rifampicin rectangle | 597 | 0.85% | 2.21% | 0.32% | 18px | 4.5% | 71px | 17.75% | 36px | 9.07% | 16px | 4% | 83px | 20.75% |
Mecillinam rectangle | 484 | 0.66% | 1.86% | 0.26% | 19px | 4.75% | 66px | 16.5% | 33px | 8.2% | 18px | 4.5% | 65px | 16.25% |
MP265 rectangle | 410 | 0.59% | 1.55% | 0.28% | 18px | 4.5% | 58px | 14.5% | 31px | 7.7% | 17px | 4.25% | 64px | 16% |
Vesicle rectangle | 393 | 0.12% | 0.26% | 0.05% | 8px | 2% | 22px | 5.5% | 14px | 3.42% | 9px | 2.25% | 21px | 5.25% |
CAM rectangle | 374 | 0.81% | 2.36% | 0.07% | 10px | 2.5% | 86px | 21.5% | 37px | 9.17% | 10px | 2.5% | 83px | 20.75% |
Control rectangle | 308 | 1.24% | 3.26% | 0.42% | 19px | 4.75% | 101px | 25.25% | 48px | 11.93% | 18px | 4.5% | 96px | 24% |
Nalidixate rectangle | 209 | 1.43% | 3.49% | 0.46% | 21px | 5.25% | 89px | 22.25% | 50px | 12.59% | 16px | 4% | 122px | 30.5% |
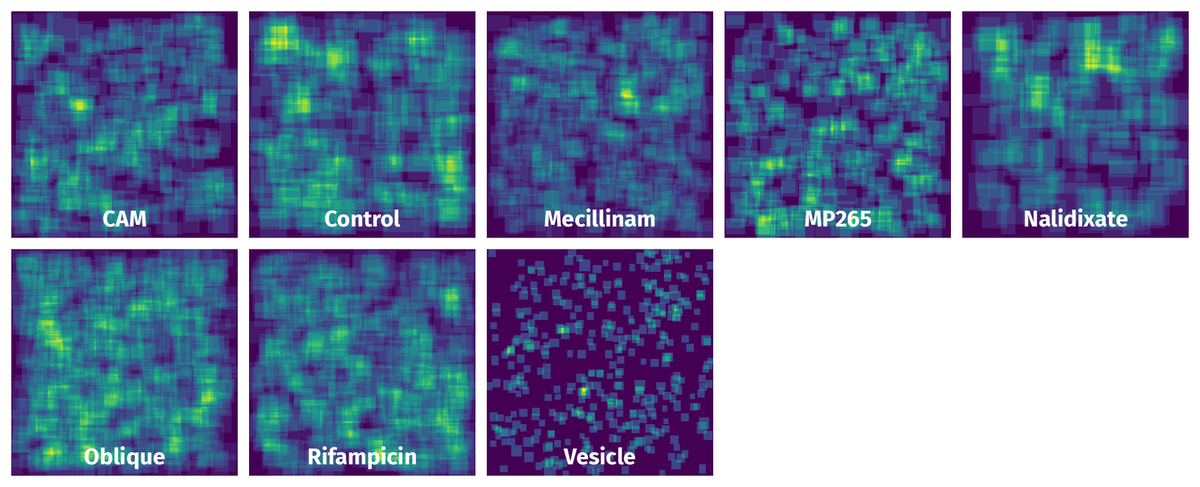
Spatial Heatmap #
The heatmaps below give the spatial distributions of all objects for every class. These visualizations provide insights into the most probable and rare object locations on the image. It helps analyze objects' placements in a dataset.
Objects #
Table contains all 3564 objects. Click a row to preview an image with annotations, and use search or pagination to navigate. Sort columns to find outliers in the dataset.
Object ID ㅤ | Class ㅤ | Image name click row to open | Image size height x width | Height ㅤ | Height ㅤ | Width ㅤ | Width ㅤ | Area ㅤ |
|---|---|---|---|---|---|---|---|---|
1➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 21px | 5.25% | 37px | 9.25% | 0.49% |
2➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 33px | 8.25% | 46px | 11.5% | 0.95% |
3➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 35px | 8.75% | 38px | 9.5% | 0.83% |
4➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 21px | 5.25% | 63px | 15.75% | 0.83% |
5➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 42px | 10.5% | 38px | 9.5% | 1% |
6➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 41px | 10.25% | 29px | 7.25% | 0.74% |
7➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 39px | 9.75% | 46px | 11.5% | 1.12% |
8➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 26px | 6.5% | 34px | 8.5% | 0.55% |
9➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 44px | 11% | 37px | 9.25% | 1.02% |
10➔ | Oblique rectangle | #6_translated_control_1.png | 400 x 400 | 46px | 11.5% | 22px | 5.5% | 0.63% |
License #
Citation #
If you make use of the DeepBacs data, please cite the following reference:
@dataset{spahn_christoph_2021_5551057,
author = {Spahn, Christoph and
Heilemann, Mike},
title = {{DeepBacs – Escherichia coli antibiotic phenotyping
object detection dataset and YOLOv2 model}},
month = oct,
year = 2021,
publisher = {Zenodo},
doi = {10.5281/zenodo.5551057},
url = {https://doi.org/10.5281/zenodo.5551057}
}
If you are happy with Dataset Ninja and use provided visualizations and tools in your work, please cite us:
@misc{ visualization-tools-for-deepbacs-dataset,
title = { Visualization Tools for DeepBacs E. Coli Dataset },
type = { Computer Vision Tools },
author = { Dataset Ninja },
howpublished = { \url{ https://datasetninja.com/deepbacs } },
url = { https://datasetninja.com/deepbacs },
journal = { Dataset Ninja },
publisher = { Dataset Ninja },
year = { 2026 },
month = { feb },
note = { visited on 2026-02-20 },
}Download #
Dataset DeepBacs E. Coli can be downloaded in Supervisely format:
As an alternative, it can be downloaded with dataset-tools package:
pip install --upgrade dataset-tools
… using following python code:
import dataset_tools as dtools
dtools.download(dataset='DeepBacs E. Coli', dst_dir='~/dataset-ninja/')
Make sure not to overlook the python code example available on the Supervisely Developer Portal. It will give you a clear idea of how to effortlessly work with the downloaded dataset.
The data in original format can be downloaded here:
Disclaimer #
Our gal from the legal dep told us we need to post this:
Dataset Ninja provides visualizations and statistics for some datasets that can be found online and can be downloaded by general audience. Dataset Ninja is not a dataset hosting platform and can only be used for informational purposes. The platform does not claim any rights for the original content, including images, videos, annotations and descriptions. Joint publishing is prohibited.
You take full responsibility when you use datasets presented at Dataset Ninja, as well as other information, including visualizations and statistics we provide. You are in charge of compliance with any dataset license and all other permissions. You are required to navigate datasets homepage and make sure that you can use it. In case of any questions, get in touch with us at hello@datasetninja.com.


