Introduction #
The Malaria Segmentation: A Dataset for Segmentation of Malaria Infected Cells consists of Giemsa stained images derived from blood samples of 16 patients infected with Plasmodium falciparum. Skilled experts meticulously annotated the different types of parasites within each image. The authors adopted a two-step methodology, which began with image segmentation and was followed by life stage classification. During the segmentation phase, each pixel was categorized as either a parasite or a non-parasite pixel using a random forest classifier. The performance of this segmentation process was assessed using metrics like classification accuracy, Dice coefficient, and free-response receiver operating characteristic (FROC) analysis. For the life stage classification, the segmented objects were divided into 8 classes, covering a range of parasite life stages, as well as other elements such as white blood cells or debris.
The dataset was derived from patient samples in Gambia. Plasmodium falciparum parasites were cultured ex-vivo for 24 to 48 hours. Giemsa-stained images revealed the parasites as dark purple entities, contrasting with the light pink appearance of red blood cells. Each image consisted of 4 by 4 sub-images, which were separated for individual analysis. The dataset comprised a total of 837 images obtained from 16 patients, with one patient omitted due to flawed staining.
To establish ground truth image segmentation, the authors manually selected and cropped images around dark-stained blobs containing parasites, white blood cells, or debris. Initial segmentation of parasites was performed using Otsu thresholding on the green channel, followed by manual adjustments and fine-tuning. Morphological operations such as imfill() and imdilate() in Matlab were employed for further refinement. The segmented cropped images were combined into a ground truth binary image.
The parasite life stages were described as follows. R = light ring, LR/ET = fat rings with dark cytoplasm, very small trophozoites with pigment; rif stage, MT = pigment, vacuole, mid-size, LT = two nuclei, very large, Esch = more than two nuclei, dark staining, very large, Lsch = still has RBC, but clear merozoites, Seg = no RBC (or very faint outline); bunch of grapes. Debris is a left-over category for those blobs that could not be clearly classified as parasites or WBCs. There were a total of 2911 parasites in different life stages, with relatively limited class imbalance. While the focus was not on WBC or debris classification, their presence was taken into account within the methodology.
Summary #
Plasmodium Falciparum from Images of Giemsa for Malaria Detection is a dataset for instance segmentation, semantic segmentation, and object detection tasks. It is used in the medical industry.
The dataset consists of 883 images with 3572 labeled objects belonging to 9 different classes including Debris, Esch, R, and other: Lsch, LT, MT, LR-ET, Seg, and WBC.
Images in the Plasmodium Falciparum from Images of Giemsa for Malaria Detection dataset have pixel-level instance segmentation annotations. Due to the nature of the instance segmentation task, it can be automatically transformed into a semantic segmentation (only one mask for every class) or object detection (bounding boxes for every object) tasks. All images are labeled (i.e. with annotations). There are no pre-defined train/val/test splits in the dataset. The dataset was released in 2019 by the Radboud University Nijmegen, Netherlands and Max Planck Institute for Developmental Biology, Germany.
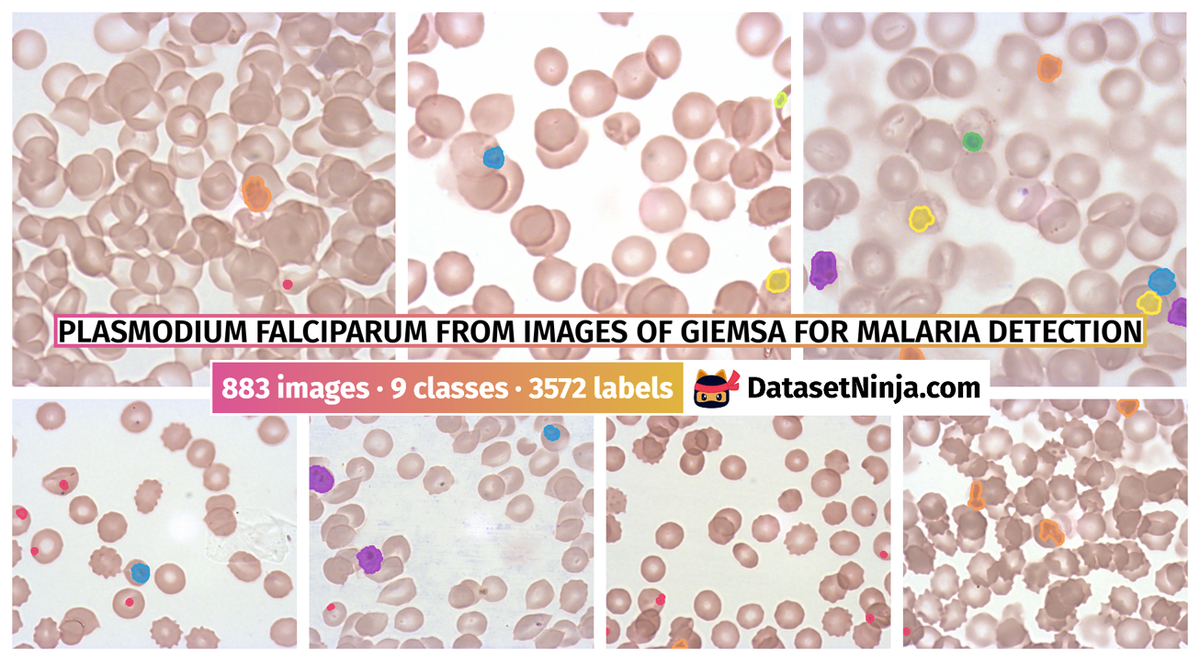
Explore #
Plasmodium Falciparum from Images of Giemsa for Malaria Detection dataset has 883 images. Click on one of the examples below or open "Explore" tool anytime you need to view dataset images with annotations. This tool has extended visualization capabilities like zoom, translation, objects table, custom filters and more. Hover the mouse over the images to hide or show annotations.

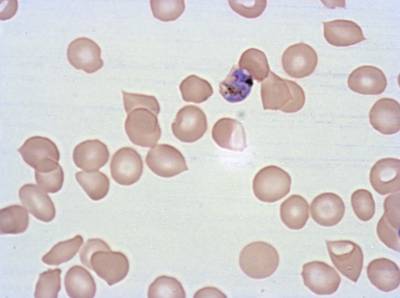

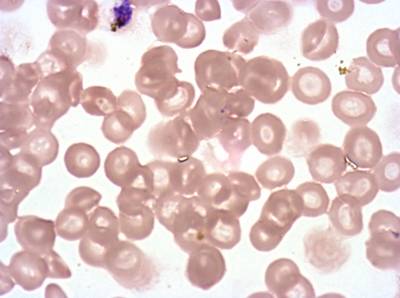

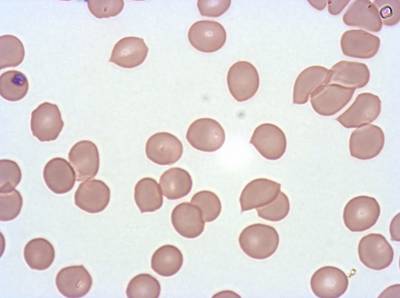

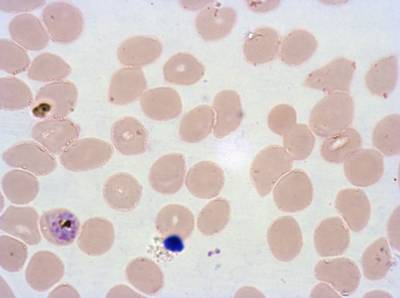

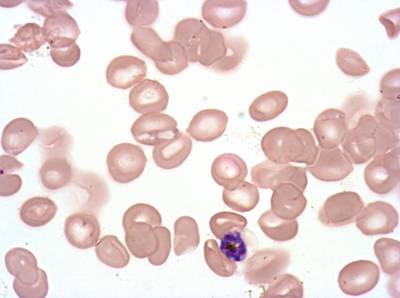

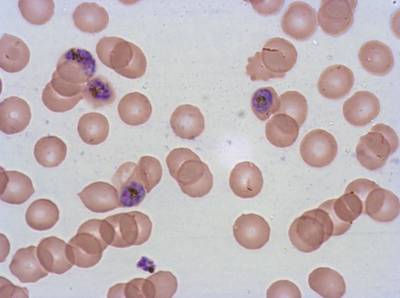

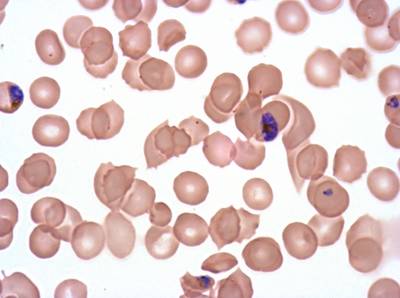

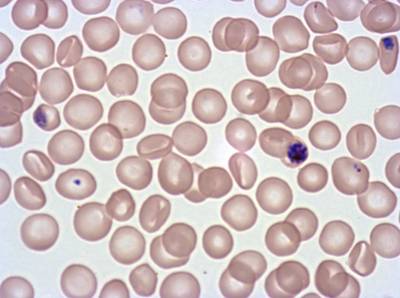

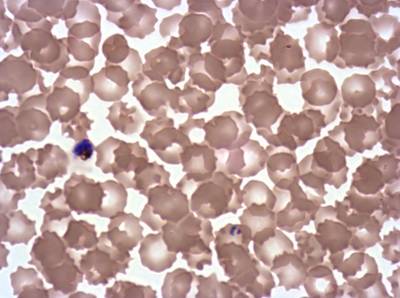

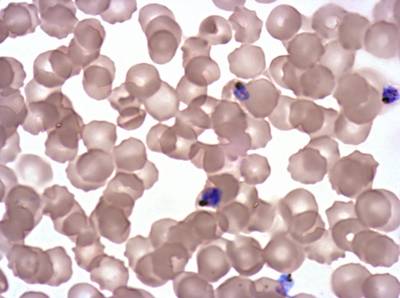

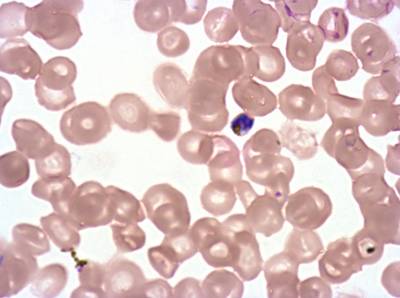

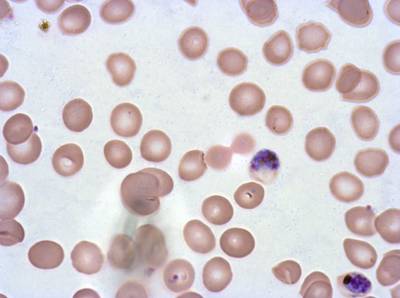

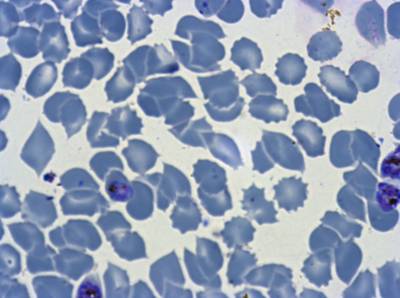

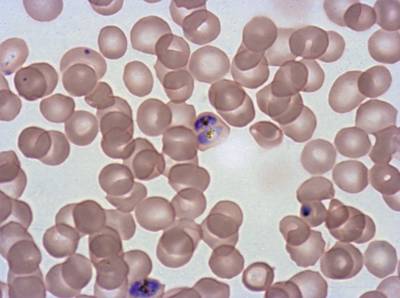

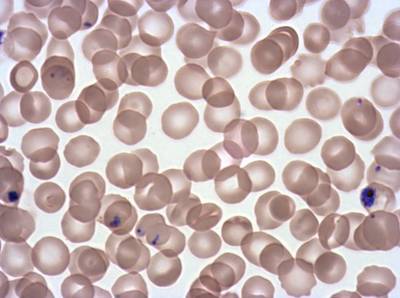

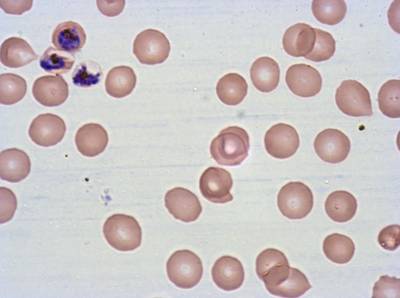

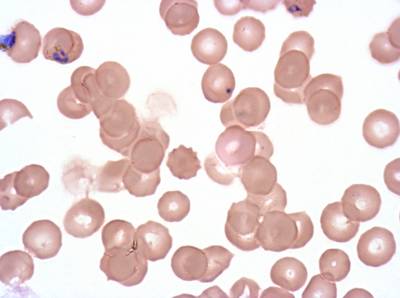

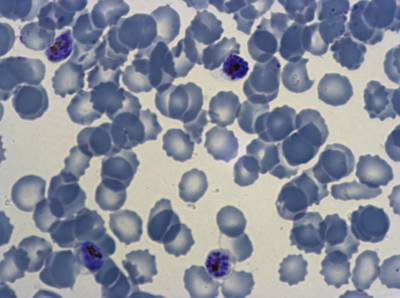

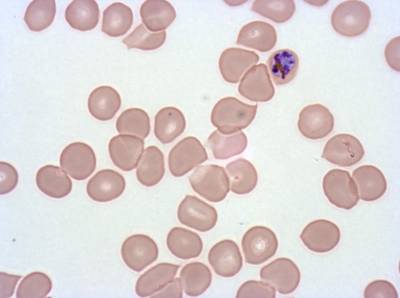





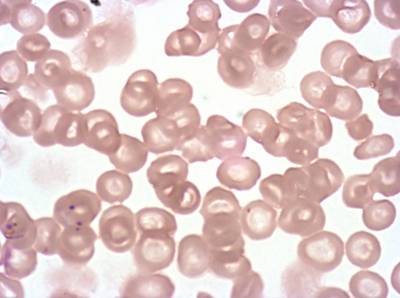

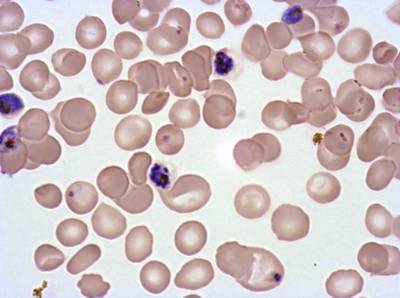

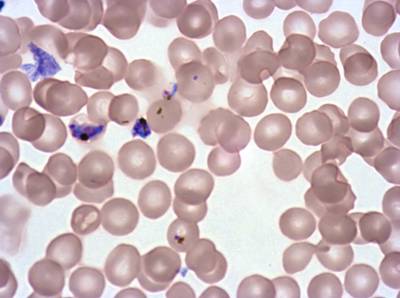

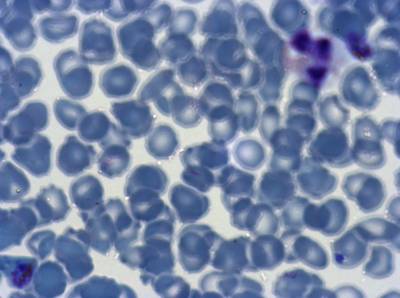

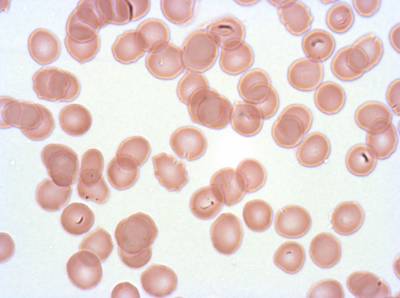

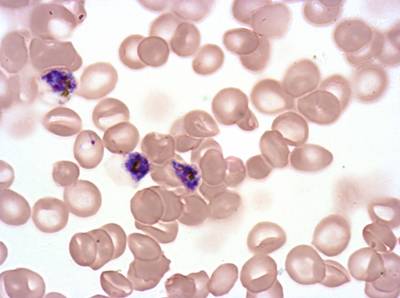

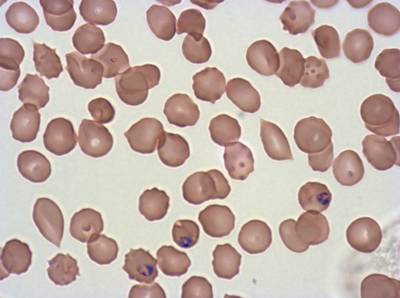
Class balance #
There are 9 annotation classes in the dataset. Find the general statistics and balances for every class in the table below. Click any row to preview images that have labels of the selected class. Sort by column to find the most rare or prevalent classes.
Class ㅤ | Images ㅤ | Objects ㅤ | Count on image average | Area on image average |
|---|---|---|---|---|
Debris➔ mask | 435 | 683 | 1.57 | 0.21% |
Esch➔ mask | 420 | 696 | 1.66 | 0.62% |
R➔ mask | 330 | 517 | 1.57 | 0.08% |
Lsch➔ mask | 307 | 439 | 1.43 | 0.62% |
LT➔ mask | 259 | 363 | 1.4 | 0.37% |
MT➔ mask | 243 | 328 | 1.35 | 0.26% |
LR-ET➔ mask | 226 | 303 | 1.34 | 0.13% |
Seg➔ mask | 148 | 200 | 1.35 | 0.6% |
WBC➔ mask | 33 | 43 | 1.3 | 1.09% |
Co-occurrence matrix #
Co-occurrence matrix is an extremely valuable tool that shows you the images for every pair of classes: how many images have objects of both classes at the same time. If you click any cell, you will see those images. We added the tooltip with an explanation for every cell for your convenience, just hover the mouse over a cell to preview the description.
Images #
Explore every single image in the dataset with respect to the number of annotations of each class it has. Click a row to preview selected image. Sort by any column to find anomalies and edge cases. Use horizontal scroll if the table has many columns for a large number of classes in the dataset.
Object distribution #
Interactive heatmap chart for every class with object distribution shows how many images are in the dataset with a certain number of objects of a specific class. Users can click cell and see the list of all corresponding images.
Class sizes #
The table below gives various size properties of objects for every class. Click a row to see the image with annotations of the selected class. Sort columns to find classes with the smallest or largest objects or understand the size differences between classes.
Class | Object count | Avg area | Max area | Min area | Min height | Min height | Max height | Max height | Avg height | Avg height | Min width | Min width | Max width | Max width |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Esch mask | 696 | 0.38% | 1.03% | 0.03% | 14px | 1.36% | 160px | 15.53% | 85px | 8.27% | 18px | 1.3% | 162px | 11.72% |
Debris mask | 683 | 0.13% | 2.15% | 0.02% | 11px | 1.07% | 219px | 21.26% | 45px | 4.39% | 17px | 1.23% | 350px | 25.33% |
R mask | 517 | 0.05% | 0.4% | 0.01% | 14px | 1.36% | 83px | 8.06% | 30px | 2.91% | 16px | 1.16% | 87px | 6.3% |
Lsch mask | 439 | 0.43% | 1.14% | 0.05% | 19px | 1.84% | 166px | 16.12% | 91px | 8.86% | 19px | 1.37% | 202px | 14.62% |
LT mask | 363 | 0.26% | 0.86% | 0.03% | 12px | 1.17% | 162px | 15.73% | 70px | 6.77% | 28px | 2.03% | 129px | 9.33% |
MT mask | 328 | 0.19% | 0.58% | 0.03% | 25px | 2.43% | 125px | 12.14% | 60px | 5.78% | 21px | 1.52% | 106px | 7.67% |
LR-ET mask | 303 | 0.1% | 0.27% | 0.02% | 18px | 1.75% | 79px | 7.67% | 42px | 4.04% | 16px | 1.16% | 88px | 6.37% |
Seg mask | 200 | 0.45% | 1.29% | 0.14% | 34px | 3.3% | 212px | 20.58% | 95px | 9.18% | 37px | 2.68% | 189px | 13.68% |
WBC mask | 43 | 0.83% | 2.38% | 0.25% | 63px | 6.12% | 219px | 21.26% | 126px | 12.23% | 41px | 2.97% | 261px | 18.89% |
Spatial Heatmap #
The heatmaps below give the spatial distributions of all objects for every class. These visualizations provide insights into the most probable and rare object locations on the image. It helps analyze objects' placements in a dataset.
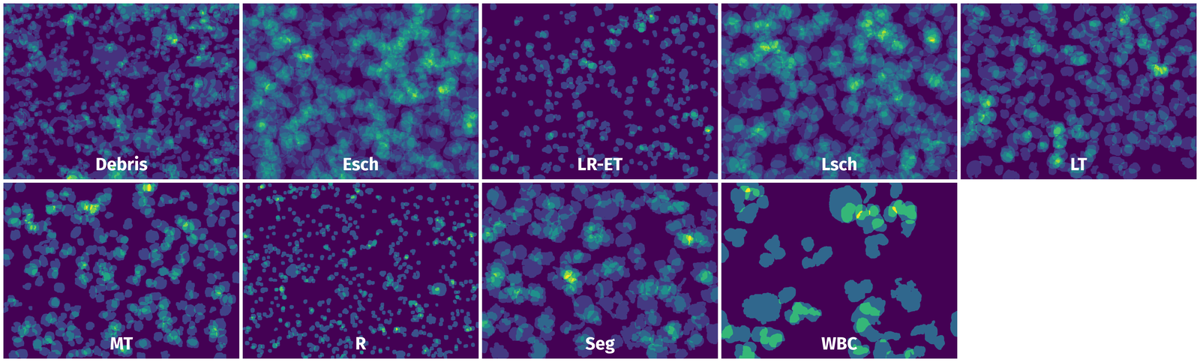
Objects #
Table contains all 3572 objects. Click a row to preview an image with annotations, and use search or pagination to navigate. Sort columns to find outliers in the dataset.
Object ID ㅤ | Class ㅤ | Image name click row to open | Image size height x width | Height ㅤ | Height ㅤ | Width ㅤ | Width ㅤ | Area ㅤ |
|---|---|---|---|---|---|---|---|---|
1➔ | LT mask | Trip 808 Day 2 08-12-05 Image 1_8.png | 1030 x 1382 | 77px | 7.48% | 97px | 7.02% | 0.39% |
2➔ | MT mask | Trip 808 Day 2 08-12-05 Image 1_8.png | 1030 x 1382 | 48px | 4.66% | 63px | 4.56% | 0.15% |
3➔ | LR-ET mask | Trip 808 Day 2 08-12-05 Image 1_8.png | 1030 x 1382 | 54px | 5.24% | 50px | 3.62% | 0.16% |
4➔ | Lsch mask | Trip 038 Day 1 01-12-05 Image 30 add_6.png | 1030 x 1382 | 126px | 12.23% | 89px | 6.44% | 0.61% |
5➔ | Esch mask | Trip 038 Day 1 01-12-05 Image 30 add_6.png | 1030 x 1382 | 96px | 9.32% | 81px | 5.86% | 0.41% |
6➔ | Esch mask | Trip 038 Day 1 01-12-05 Image 30 add_6.png | 1030 x 1382 | 108px | 10.49% | 103px | 7.45% | 0.58% |
7➔ | LR-ET mask | Trip 038 Day 1 01-12-05 Image 30 add_6.png | 1030 x 1382 | 68px | 6.6% | 42px | 3.04% | 0.15% |
8➔ | Lsch mask | Trip 804 Day 1 02-12-05 Image 1_5.png | 1030 x 1382 | 106px | 10.29% | 102px | 7.38% | 0.58% |
9➔ | MT mask | Trip 804 Day 1 02-12-05 Image 1_5.png | 1030 x 1382 | 60px | 5.83% | 49px | 3.55% | 0.13% |
10➔ | Esch mask | Trip 804 Day 1 02-12-05 Image 1_5.png | 1030 x 1382 | 94px | 9.13% | 90px | 6.51% | 0.44% |
License #
Citation #
If you make use of the Malaria Segmentation data, please cite the following reference:
Abbas, Syed Saiden; M. H. Dijkstra, Tjeerd (2019),
“Malaria-Detection-2019”,
Mendeley Data, V1, doi: 10.17632/5bf2kmwvfn.1
If you are happy with Dataset Ninja and use provided visualizations and tools in your work, please cite us:
@misc{ visualization-tools-for-malaria-segmentation-dataset,
title = { Visualization Tools for Plasmodium Falciparum from Images of Giemsa for Malaria Detection Dataset },
type = { Computer Vision Tools },
author = { Dataset Ninja },
howpublished = { \url{ https://datasetninja.com/malaria-segmentation } },
url = { https://datasetninja.com/malaria-segmentation },
journal = { Dataset Ninja },
publisher = { Dataset Ninja },
year = { 2026 },
month = { feb },
note = { visited on 2026-02-24 },
}Download #
Dataset Plasmodium Falciparum from Images of Giemsa for Malaria Detection can be downloaded in Supervisely format:
As an alternative, it can be downloaded with dataset-tools package:
pip install --upgrade dataset-tools
… using following python code:
import dataset_tools as dtools
dtools.download(dataset='Plasmodium Falciparum from Images of Giemsa for Malaria Detection', dst_dir='~/dataset-ninja/')
Make sure not to overlook the python code example available on the Supervisely Developer Portal. It will give you a clear idea of how to effortlessly work with the downloaded dataset.
The data in original format can be downloaded here.
Disclaimer #
Our gal from the legal dep told us we need to post this:
Dataset Ninja provides visualizations and statistics for some datasets that can be found online and can be downloaded by general audience. Dataset Ninja is not a dataset hosting platform and can only be used for informational purposes. The platform does not claim any rights for the original content, including images, videos, annotations and descriptions. Joint publishing is prohibited.
You take full responsibility when you use datasets presented at Dataset Ninja, as well as other information, including visualizations and statistics we provide. You are in charge of compliance with any dataset license and all other permissions. You are required to navigate datasets homepage and make sure that you can use it. In case of any questions, get in touch with us at hello@datasetninja.com.


