Introduction #
The PolypGen: A Polyp Segmentation and Detection Generalisation Dataset is one of the most comprehensive detection and pixel-level segmentation datasets curated by a team of computational scientists and expert gastroenterologists. The authors have curated a dataset from six unique centres incorporating more than 300 patients which is crucial fue to automate polyp detection and segmentation. Polyps in the colon are widely known cancer precursors identified by colonoscopy - the polyp’s number, size, and surface structure are linked to the risk of colon cancer. The dataset includes both single frame and sequence data with 3762 annotated polyp labels with precise delineation of polyp boundaries verified by six senior gastroenterologists.
PolypGen data was collected from 6 different centers. More than 300 unique patient videos/frames were used for this study. The general purpose of this diverse dataset is to allow the robust design of deep learning models and their validation to assess their generalizability capability. In this context, authors have proposed different dataset configurations for training and out-of-sample validation and proposed unique generalization assessment metrics to reveal the strength of deep learning methods. The below authors provide a comprehensive description of dataset collection, annotation strategies and their quality, ethical guidelines, and metric evaluation strategies.
| Dataset | Findings | Size | Availability |
|---|---|---|---|
| Kvasir-SEG | Polyps | 1000 images† | Open academic |
| HyperKvasir | GI findings including polyps | 110,079 images and 374 videos | Open academic |
| Kvasir-Capsule | GI findings including polyps | 4,741,504 images | Open academic |
| CVC-ColonDB | Polyps | 380 images†† | By request• |
| ETIS-Larib Polyp DB | Polyps | 196 images† | Open academic |
| EDD202015 | GI lesions including polyps | 386 images | Open academic |
| CVC-ClinicDB | Polyps | 612 images† | Open academic |
| CVC-VideoClinicDB | Polyps | 11,954 images† | By request• |
| ASU-Mayo polyp database | Polyps | 18,781 images† | By request• |
| KID | Angiectasia, bleeding, inflammations | 2371 images, 47 videos | Open academic• |
| Atlas of GI Endoscope | GI lesions | 1295 images | Unknown• |
| El salvador atlas | GI lesions | 5071 video clips | Open academic♣️ |
| PolypGen | Multi-centre colon polyps | 1537 images†& 2225 video sequence | Open academic |
Table 1. An overview of existing gastrointestinal (GI) lesion datasets including polyps: the number of images or videos
along with the availability type is provided.
| Centre | System info | Ethical approval | Patient consenting type |
|---|---|---|---|
| Ambroise Paré Hospital, Paris, France | Olympus Exera 195 | N° IDRCB: 2019-A01602-55 | Endospectral study |
| Istituto Oncologico Veneto, Padova, Italy | Olympus endoscope H190 | NA | Generic patients consent |
| Centro Riferimento Oncologico, IRCCS, Italy | Olympus VG-165, CV180, H185 | NA | Generic patients consent |
| Oslo University Hospital, Oslo, Norway | Olympus Evis Exera III, CF 190 | Exempted† | Written informed consent |
| John Radcliffe Hospital, Oxford, UK | GIF-H260Z, EVIS Lucera CV260 | REC Ref: 16/YH/0247 | Universal consent |
| University of Alexandria, Alexandria, Egypt | Olympus Exera 160AL, 180AL | NA | Written informed consent |
Table 2. Data collection information for each centre: Data acquisition system and patient consenting information.
A consortium of six different medical data centres (hospitals) was built where each data centre provided videos and image frames from at least 50 unique patients. The videos and image samples were collected and sent by the senior gastroenterologists involved in this project. The collected dataset consisted of both polyp and normal mucosa colonoscopy acquisitions. To incorporate the nature of polyp occurrences and maintain heterogeneity in the data distribution, the following protocol was adhered to for establishing the dataset:
- Single frame sampling from each patient video incorporated different viewpoints
- Sequence frame sampling consisted of both visible and invisible polyp frames (in most cases) with a minimal gap
- While single frame data consisted of all polyp instances in that patient, sequence frame data consisted of only a localised targeted polyp
- Positive sequence included both positive and negative polyp instances but from videos with confirmed polyp location while for negative sequence only patient videos with normal mucosa were used
An overview of the number of samples comprising positive samples and negative samples is presented in Fig. 2 a. The total positive samples of 3762 frames are released comprising 484, 1166, 457, 677, 458 and 520 frames from centres C1, C2, C3, C4, C5 and C6, respectively. These frames consist of 1537 single frames (1449 frames from C1-C5 also provided in EndoCV2021 challenge and 88 frames from C6), and 2225 sequence frames with the majority of sequence data sampled from centres C2 (865), C4 (450), and C6 (432). The number of polyp counts for pixel-level annotation of small (≤ 100×100), medium (between > 100 × 100 pixels and ≤ 200 × 200 pixels), large (≥ 200 × 200 pixels) sized polyps from each centre including no polyp frames but frames in close proximity of polyp are represented as histogram plot (Fig. 2 b). The total annotations for polyp that authors release is 3447. All these polyp samples are verified by expert gastroenterologists.


Figure 2. PolypGen dataset: (a) Positive (both single and sequence frames) and negative samples (sequence only) from each centre, and (b) polyp size-based histogram plot for positive samples showing variable-sized annotated polyps in the dataset (small is ≤ 100×100 pixels; medium is > 100×100 ≤ 200×200, and large is > 200×200 pixels). Null represents no polyp present in the sample.
The authors have provided both still image frames and continuous short video sequence data with their corresponding annotations. The positive and negative samples in the dataset of the polyp generalisation (PolypGen) are further detailed below.

Figure 3. Sample polyp annotations from each centre: Segmentation area with boundaries and corresponding bounding box/boxes overlaid images from all six centres. Samples include both small-sized polyp (< 10000 pixels) including some flat polyp samples to large-sized (≥ 40000 pixels) polyps and polyps during the resection procedure such as polyps with blue dyes.
Positive samples
Positive samples consist of video frames from the patient with a diagnosed polyp case. The selected frames may or may not have the polyp in them but may be located near the chosen frame. Nevertheless, a majority of these frames consist of at least one polyp in the frame. For the sequence positive samples, the continuity of the appearance and disappearance of the polyp similar to the real scenario has been taken into account and thus these frames can have a mixture of polyp instances and frames with normal mucosa. Table 3 is provided to detail the characteristics of 23 sequence data included in our dataset. It can be observed from Figure 4 that varied-sized polyps are included in the dataset with variable viewpoints, occlusions and instruments. Exemplary pixel-level annotations of positive polyp samples for each centre and their corresponding bounding boxes are presented in Fig. 3.
| Sequence | Description | Artifact |
|---|---|---|
| seq1 | Normal mucosa | Light reflections; green patch |
| seq2 | 5 mm polyp at 6 o’clock | Partially covered with stool; reflections; green patch |
| seq3 | Polyp at distance, 4 o’clock | Light reflection from liquid; green patch |
| seq4 | 2-3 mm polyp | Liquid covering half of the image; green patch |
| seq5 | 5 mm polyp catched by a snare | Partial occlusion by biopsy instrument |
| seq6 | Polyp covering half of the circumference | Cap; green patch |
| seq7 | Normal mucosa | Light reflection; some remnant stool; green patch |
| seq8 | Typical flat cancer | Light reflection; green patch |
| seq9 | 2 mm polyp at 2 o’clock | Light reflection; green patch |
| seq10 | Subtle small protrusions | Some remnant stool |
| seq11 | Polyp at 2-3 o’clock | Light reflections in the periphery |
| seq12 | Dye lifted 4-5 mm polyp | Low contrast |
| seq13 | 6-7 mm polyp catched with a snare | Low contrast; small reflections |
| seq14 | Paris 1 p polyp, large long stalk, JNET 2a | Lifted by Indigo Carmine, snare placed around the stalk |
| seq15 | Paris 1 s JNET2a polyp and 1 Paris 1 sp to the left | Lifted by Indigo Carmine |
| seq16 | Paris 1 p polyp, large long stalk, JNET 2a | Lifted by Indigo Carmine |
| seq17 | Paris 1 sp polyp | Light reflections make surface assessment impossible |
| seq18 | Difficult interpretation | Blurry image and reduced view |
| seq19 | Paris 1 p polyp, large long stalk, JNET 2a | Less contrast and slightly occluded |
| seq20 | Half of the polyp visible | Blurry image, with some blood on the mucosa |
| seq21 | Two adenomas polyp | Blurry image |
| seq22 | Adenomas polyp | Blurry image makes exact diagnosis impossible |
| seq23 | Serrated polyp | Perfect clean mucosa, minor light reflections |
Table 3. Positive sample sequence summarised attribute: Total of 23 sequences are provided as positive sample sequences for patients with polyp instances during colonoscopy examination. Here JNET refers to the Japan NBI Expert Team classification score. These sequences depict different-sized polyps and locations with different artefacts and varying visibility. Sequences referring to one selected image are shown in Fig. 4.
Negative samples
Negative samples mostly refer to the negative sequences released in this dataset, i.e. no polyp frames. These sequences are taken from patient videos which consisted of confirmed absence of polyps (i.e., normal mucosa) in the acquired videos or at areas away from the polyp occurrences. It includes cases with anatomies such as colon linings, light reflections and mucosa covered with stool that may be confused with polyps (see Figure 5 and corresponding negative sequence attributes in Table 4).
Annotation strategies and quality assurance
A team of 6 senior gastroenterologists (with over 20 years of experience in endoscopy), two experienced post-doctoral researchers, and one PhD student were involved in the data collection, data sorting, annotation and the review process of the quality of annotations. For details on data collection and data sorting please refer to Section Video acquisition, collection and dataset construction. All annotations were performed by a team of three experienced researchers using an online annotation tool called Labelbox. The dataset was divided equally between the three reviewers for the annotation process where each researcher annotated a specific group of frames. However, all the annotated frames were revised by the senior gastroenterologists’ team. Each annotation was later cross-validated for accurate segmentation margins by the team and by the centre expert. Further, an independent binary review process was then assigned to a senior gastroenterologist, in most cases experts from different centres were assigned. A protocol for manual annotation of polyp was designed to minimise the heterogeneity in the manual delineation process. The protocol was in detail discussed with the clinical experts and the annotators during several weekly meetings. Here, the authors only present a brief on the important aspects of the annotation that should be taken care of during annotations. Example samples were provided by expert endoscopists to the annotators especially this was the case in the video annotations. The set protocols are listed below (refer to Fig. 3 for final ground truth annotations):

Figure 4. Positive sequence data: Representative samples chosen from 23 sequences of the provided positive samples data.
A summary description is provided in Table 3. Parts of the images have been cropped for visualization.
- Clear raised polyps: Boundary pixels should include only protruded regions. Precaution has to be taken when delineating along the normal colon folds
- Inked polyp regions: Only part of the non-inked appearing object delineation
- Polyps with instrument parts: Annotation should not include instrument and is required to be carefully delineated and may form more than one object
- Pedunculated polyps: Annotation should include all raised regions unless appearing on the fold
- Flat polyps: Zooming the regions identified with flat polyps before manual delineation. Also, consulting centre expert if needed.

Figure 5. Negative sequence data: Representative samples chosen from each sequence of the provided negative sample data. A summary description is provided in Table 4. Parts of the images have been cropped for visualization.
- Video sequence annotation: One sample from an expert gastroenterologist was provided for sequences that showed difficulty in distinguishing between mucosa and polyp. Polyps that are distant and not clearly visible were also not annotated as polyps.
- Tackling with occlusion: Polyps that were occluded with stool or instrument were required to exclude the parts of mucosa that were obstructed.
- Cancerous mucosa: Mucosa that were already cancerous but did not appear as polyps were excluded from the annotation. However, a raised mucosal surface that characterised adenomatous polyps was included.
Each of these annotated masks was reviewed by expert gastroenterologists. During this review process, a binary score was provided by the experts depending on whether the annotations were clinically acceptable or not. Some of the experts also provided feedback on the annotation and these images were placed into ambiguous categories for further rectification based on expert feedback. This ambiguous category was then jointly annotated by two researchers and further sent for review to one expert. The outcome of these quality checks is provided in Figure 6. It can be observed that a large fraction (30.5%) of annotations were rejected (excluding ambiguous batches, the total annotations were 2213, among which only 1537 were accepted and 676 frames were rejected). Similarly, the ambiguous batch that included correction of annotations after the first review also recorded 34.17% of rejected frames on the second review.
| Sequence | Description | Artifact |
|---|---|---|
| seq1_neg | Normal vascular pattern | Light reflections in the periphery; not clean lens |
| seq2_neg | Normal vascular pattern | Contracted bowel; light reflections |
| seq3_neg | Mucosa not satisfactorily visualized | Stool covers the field of view |
| seq4_neg | Reduced vascular pattern | Light reflections and a small amount of stool |
| seq5_neg | Reduced vascular pattern | Light reflections |
| seq6_neg | Normal vascular pattern | Light reflections; biopsy forceps |
| seq7_neg | Normal vascular pattern | Very close to the luminal wall |
| seq8_neg | Normal vascular pattern | Blurry; semi-opaque liquid; cap |
| seq9_neg | Normal vascular pattern | Blurry; semi-opaque liquid; cap |
| seq10_neg | Not possible to assess the mucosa | Blurry; occluded |
| seq11_neg | Normal vascular pattern | Light reflections in the periphery; bubble on the lens |
| seq12_neg | Normal vascular pattern | Not a clean lens, mucosa covered by stool |
| seq13_neg | Probably normal vascular pattern; Not possible to assess the mucosa | Air bubbles; remnant stool; too close to the mucosa, blur, reflections |
| seq14_neg | Clean bowel, normal vascular pattern | Very close to the mucosa in all |
| seq15_neg | Clean bowel, normal vascular pattern | Some bubbles and light reflections |
| seq16_neg | Clean bowel, normal vascular pattern | Some bubbles and light reflection |
| seq17_neg | Clean bowel, normal vascular pattern | Very close to the mucosa in all |
| seq18_neg | Clean bowel, normal vascular pattern, well distended | Some stool residues |
| seq19_neg | Clean bowel, normal vascular pattern, well distended | Some liquid residues |
| seq20_neg | Clean bowel, normal vascular pattern, well distended | Some stool residues and reflections |
| seq21_neg | Clean bowel, normal vascular pattern | Very close, minor stool residues in last images |
| seq22_neg | Clean bowel, normal vascular pattern, well distended | Some liquid and stool residues, reflections |
| seq23_neg | Perfect clean bowel, normal vascular pattern, well distended | Some light reflections |
Table 4. Negative sample sequence summarised attribute: Total of 23 sequences are provided as negative sample sequences for patients with no polyp during colonoscopy examination. These sequences depict different artefacts and varying visibility of vascular patterns and occlusion of the mucosa.

Figure 6. Annotation quality review: Total curated frames along with accepted and rejected frame numbers during annotation quality review by experts for single-frame data. Annotated frames with % of flat and protruded polyps categorised during annotation are also provided
A subset of this dataset (from C1 - C5 except C6) forms the dataset of our EndoCV2021 challenge (Addressing generalisability in polyp detection and segmentation) training data, i.e., an event held in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI 2021), Nice, France. The current released data consists of additional positive and negative frames for both single and sequence data and a 6th centre data (C6). The presented version does not consist of training and test splits and users are free to apply their own strategies as applicable to the nature of their work. To access the complete dataset, users are requested to create a Synapse account and then the compiled dataset can be downloaded at source which has been published under Creative Commons 4.0 International (CC BY) licence. The dataset can only be used for educational and research purposes and must cite this paper. All collected data has been obtained through written patient consent or through ethical approval as tabulated in Table 2.
Summary #
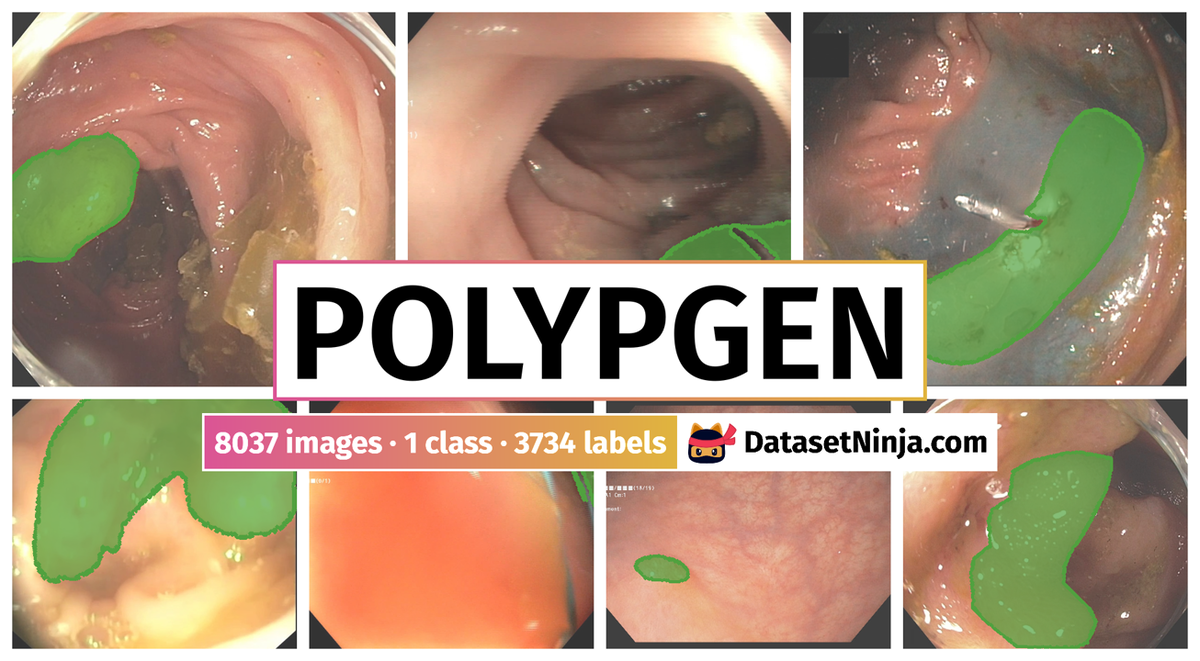
PolypGen: A Polyp Segmentation and Detection Generalisation Dataset from EndoCV2021 Challenge is a dataset for instance segmentation, semantic segmentation, object detection, and classification tasks. It is used in the medical industry.
The dataset consists of 8037 images with 3734 labeled objects belonging to 1 single class (polyp).
Images in the PolypGen dataset have pixel-level instance segmentation annotations. Due to the nature of the instance segmentation task, it can be automatically transformed into a semantic segmentation (only one mask for every class) or object detection (bounding boxes for every object) tasks. There are 4916 (61% of the total) unlabeled images (i.e. without annotations). There are no pre-defined train/val/test splits in the dataset. Alternatively, the dataset could be split into 2 cases: negative (4275 images) and positive (3762 images). Also institute and sequence tags are included. The dataset was released in 2022 by the UK-NOR-EGY-IT-SWE-FR-US joint research group.
Explore #
PolypGen dataset has 8037 images. Click on one of the examples below or open "Explore" tool anytime you need to view dataset images with annotations. This tool has extended visualization capabilities like zoom, translation, objects table, custom filters and more. Hover the mouse over the images to hide or show annotations.

































Class balance #
There are 1 annotation classes in the dataset. Find the general statistics and balances for every class in the table below. Click any row to preview images that have labels of the selected class. Sort by column to find the most rare or prevalent classes.
Class ㅤ | Images ㅤ | Objects ㅤ | Count on image average | Area on image average |
|---|---|---|---|---|
polyp➔ mask | 3121 | 3734 | 1.2 | 10.11% |
Images #
Explore every single image in the dataset with respect to the number of annotations of each class it has. Click a row to preview selected image. Sort by any column to find anomalies and edge cases. Use horizontal scroll if the table has many columns for a large number of classes in the dataset.
Object distribution #
Interactive heatmap chart for every class with object distribution shows how many images are in the dataset with a certain number of objects of a specific class. Users can click cell and see the list of all corresponding images.
Class sizes #
The table below gives various size properties of objects for every class. Click a row to see the image with annotations of the selected class. Sort columns to find classes with the smallest or largest objects or understand the size differences between classes.
Class | Object count | Avg area | Max area | Min area | Min height | Min height | Max height | Max height | Avg height | Avg height | Min width | Min width | Max width | Max width |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
polyp mask | 3734 | 8.45% | 80.91% | 0% | 1px | 0.09% | 1080px | 100% | 330px | 33.07% | 1px | 0.07% | 1716px | 100% |
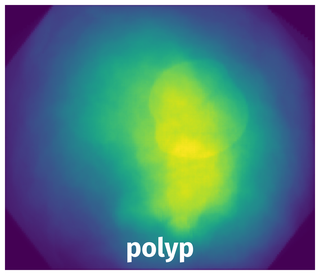
Spatial Heatmap #
The heatmaps below give the spatial distributions of all objects for every class. These visualizations provide insights into the most probable and rare object locations on the image. It helps analyze objects' placements in a dataset.
Objects #
Table contains all 3734 objects. Click a row to preview an image with annotations, and use search or pagination to navigate. Sort columns to find outliers in the dataset.
Object ID ㅤ | Class ㅤ | Image name click row to open | Image size height x width | Height ㅤ | Height ㅤ | Width ㅤ | Width ㅤ | Area ㅤ |
|---|---|---|---|---|---|---|---|---|
1➔ | polyp mask | C1_962OLCV1_100H0004.jpg | 1080 x 1350 | 307px | 28.43% | 369px | 27.33% | 1.94% |
2➔ | polyp mask | C3_C3_EndoCV2021_00464.jpg | 1080 x 1350 | 102px | 9.44% | 241px | 17.85% | 0.99% |
3➔ | polyp mask | seq9_7_endocv2021_positive_228.jpg | 1080 x 1350 | 346px | 32.04% | 139px | 10.3% | 2.39% |
4➔ | polyp mask | seq5_EndoCV2021_seq5_236.jpg | 1064 x 1350 | 329px | 30.92% | 377px | 27.93% | 6.07% |
5➔ | polyp mask | seq13_EndoCV2021_seq13_788.jpg | 1064 x 1350 | 902px | 84.77% | 568px | 42.07% | 20.32% |
6➔ | polyp mask | C3_C3_EndoCV2021_00429.jpg | 1080 x 1350 | 741px | 68.61% | 783px | 58% | 29.03% |
7➔ | polyp mask | seq12_EndoCV2021_seq12_476.jpg | 1010 x 900 | 97px | 9.6% | 96px | 10.67% | 0.69% |
8➔ | polyp mask | seq21_seq6_C6_396.jpg | 710 x 910 | 118px | 16.62% | 147px | 16.15% | 1.59% |
9➔ | polyp mask | seq21_seq6_C6_396.jpg | 710 x 910 | 193px | 27.18% | 432px | 47.47% | 9.74% |
10➔ | polyp mask | seq22_seq7_C6_390.jpg | 700 x 830 | 174px | 24.86% | 193px | 23.25% | 3.87% |
License #
Citation #
If you make use of the PolypGen data, please cite the following reference:
[1] Ali, Sharib, Jha, Debesh, Ghatwary, Noha et al. (2021) PolypGen: A
multi-center polyp detection and segmentation dataset for generalisability
assessment. arXiv.
[2] Ali, Sharib, et al. "Assessing generalisability of deep learning-based polyp
detection and segmentation methods through a computer vision challenge." arXiv
preprint arXiv:2202.12031 (2022).
[3] Ali S, Dmitrieva M, Ghatwary N, Bano S, Polat G, Temizel A, et al. Deep
learning for detection and segmentation of artefact and disease instances in
gastrointestinal endoscopy. Medical Image Analysis. 2021:102002.
If you are happy with Dataset Ninja and use provided visualizations and tools in your work, please cite us:
@misc{ visualization-tools-for-polypgen-dataset,
title = { Visualization Tools for PolypGen Dataset },
type = { Computer Vision Tools },
author = { Dataset Ninja },
howpublished = { \url{ https://datasetninja.com/polypgen } },
url = { https://datasetninja.com/polypgen },
journal = { Dataset Ninja },
publisher = { Dataset Ninja },
year = { 2025 },
month = { sep },
note = { visited on 2025-09-14 },
}Download #
Dataset PolypGen can be downloaded in Supervisely format:
As an alternative, it can be downloaded with dataset-tools package:
pip install --upgrade dataset-tools
… using following python code:
import dataset_tools as dtools
dtools.download(dataset='PolypGen', dst_dir='~/dataset-ninja/')
Make sure not to overlook the python code example available on the Supervisely Developer Portal. It will give you a clear idea of how to effortlessly work with the downloaded dataset.
Disclaimer #
Our gal from the legal dep told us we need to post this:
Dataset Ninja provides visualizations and statistics for some datasets that can be found online and can be downloaded by general audience. Dataset Ninja is not a dataset hosting platform and can only be used for informational purposes. The platform does not claim any rights for the original content, including images, videos, annotations and descriptions. Joint publishing is prohibited.
You take full responsibility when you use datasets presented at Dataset Ninja, as well as other information, including visualizations and statistics we provide. You are in charge of compliance with any dataset license and all other permissions. You are required to navigate datasets homepage and make sure that you can use it. In case of any questions, get in touch with us at hello@datasetninja.com.


