Introduction #
The authors of the Dataset of Laryngeal Endoscopic Images for Semantic Segmentation assessed existing segmentation methods. In medical image analysis, the automated segmentation of anatomical structures is crucial for autonomous diagnosis and various computer-aided interventions, including those involving robots. The examination of laryngeal endoscopic images holds the promise of early pathology detection. Given that vocal folds, the primary functional organ within the larynx, are delicate structures critical for surgery, computer vision can play a pivotal role in assisting physicians in preserving or restoring voice functionality. This synergy involves combining augmented reality, robotics, laser surgery, and image processing methods. Notably, the segmentation of laryngeal images stands out as a key component for the effective implementation of such a comprehensive system.
Dataset description
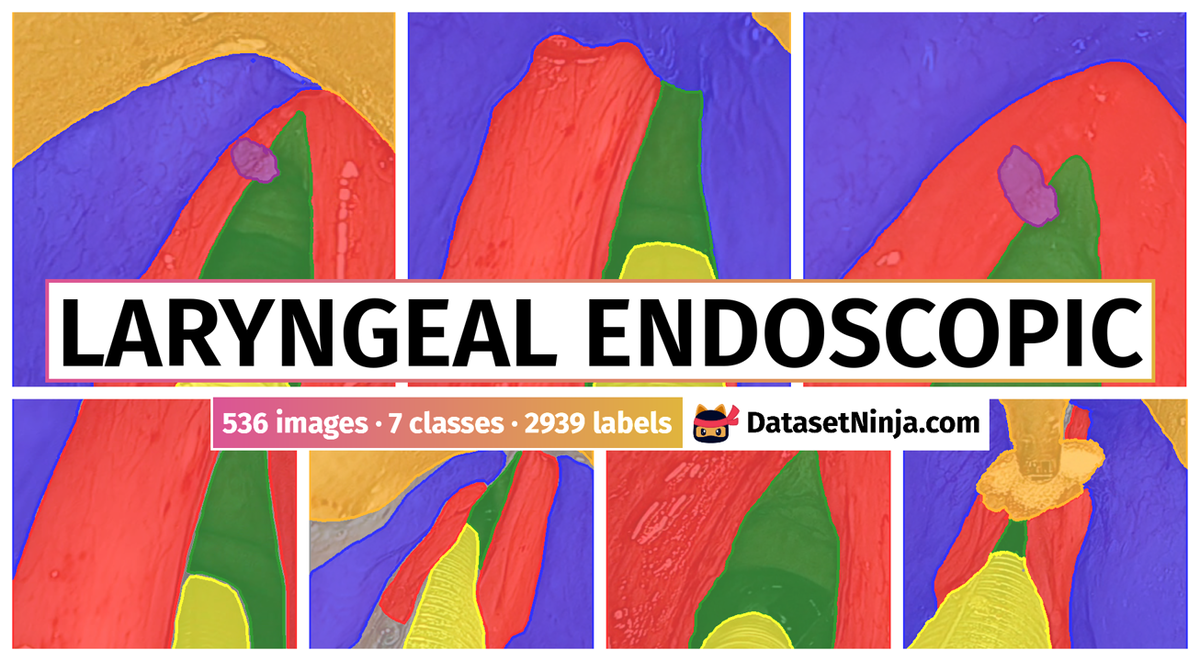
The dataset contain 536 manually segmented color images of the larynx during two different resection surgeries with a resolution of 512×512 pixels. The images have been captured with a stereo endoscope (VSii, Visionsense, Petach-Tikva, Israel). They are categorized in the 7 different classes: void, vocal folds, other tissue, glottal space, pathology, surgical tool.
First row: Examples from vocal folds dataset. Second row: Manually segmented ground truth label maps with classes vocal folds (red), other tissue (blue), glottal space (green), pathology (purple), surgical tool (orange), intubation (yellow) and void (gray).
The dataset consists of 5 different sequences from two patients. The sequences have following characteristics:
- SEQ1: pre-operative with clearly visible tumor on vocal fold, changes in translation, rotation, scale, no instruments visible, without intubation
- SEQ2: pre-operative with clearly visible tumor, visible instruments, changes in translation and scale, with intubation
- SEQ3–4: post-operative with removed tumor, damaged tissue, changes in translation and scale, with intubation
- SEQ5–7: pre-operative with instruments manipulating and grasping the vocal folds, changes in translation and scale, with intubation
- SEQ8: post-operative with blood on vocal folds, instruments and surgical dressing, with intubation
Subsequent images have a temporal contiguity as they are sampled uniformly from videos. To reduce inter-frame correlation, images were extracted from the original videos only once per second. In the comparative study SEQ4–SEQ6 were not used due to high similarity to SEQ3 and SEQ7 respectively, as they do not offer any additional variance to the dataset. Segmentations have been manually created on a pen display (DTK-2241, K. K. Wacom).
Number of annotated pixels per class in the dataset.
Summary #
A Dataset of Laryngeal Endoscopic Images for Semantic Segmentation is a dataset for a semantic segmentation task. It is used in the medical and robotics industries.
The dataset consists of 536 images with 2939 labeled objects belonging to 7 different classes including vocal folds, void, glottal space, and other: other tissue, intubation, surgical tool, and pathology.
Images in the Laryngeal Endoscopic dataset have . All images are labeled (i.e. with annotations). There are no pre-defined train/val/test splits in the dataset. Alternatively, the dataset could be split into 6 sequences: seq8 (186 images), seq7 (122 images), seq4 (66 images), seq6 (44 images), seq3 (26 images), and seq5 (26 images). Additionally, every image marked with the patient tag (1 or 2). The dataset was released in 2020 by the Institute of Mechatronic Systems, Hannover, Germany.
Explore #
Laryngeal Endoscopic dataset has 536 images. Click on one of the examples below or open "Explore" tool anytime you need to view dataset images with annotations. This tool has extended visualization capabilities like zoom, translation, objects table, custom filters and more. Hover the mouse over the images to hide or show annotations.




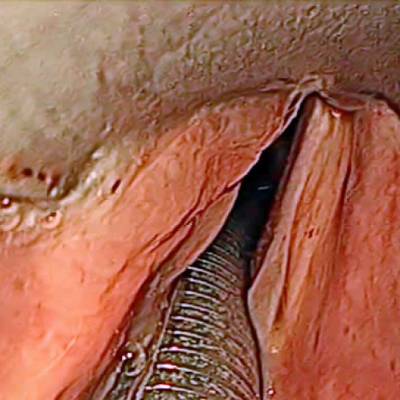



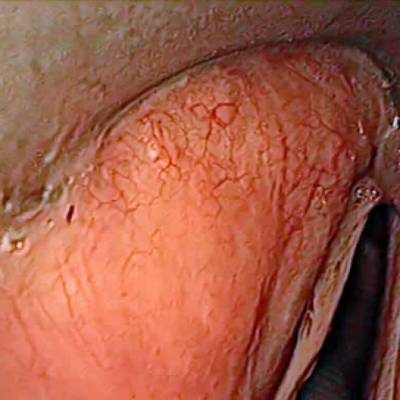

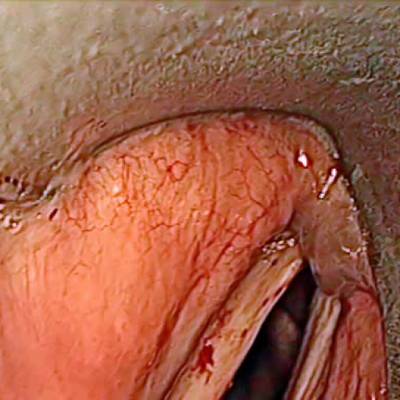

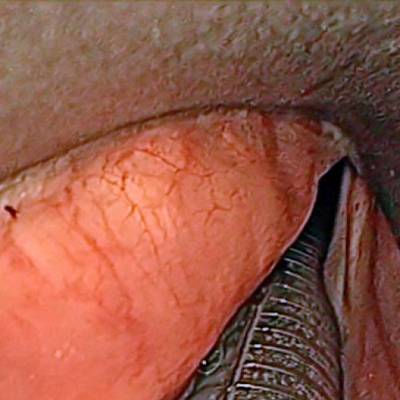

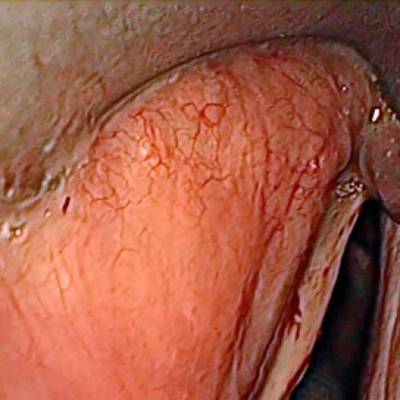

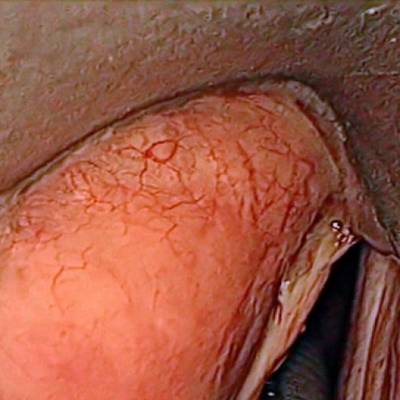

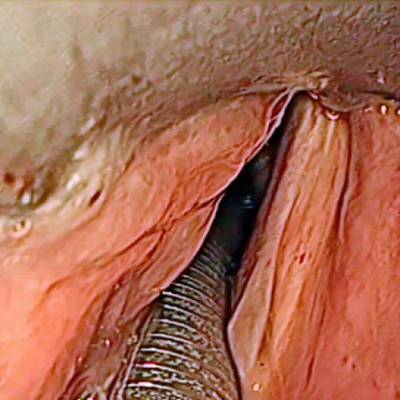

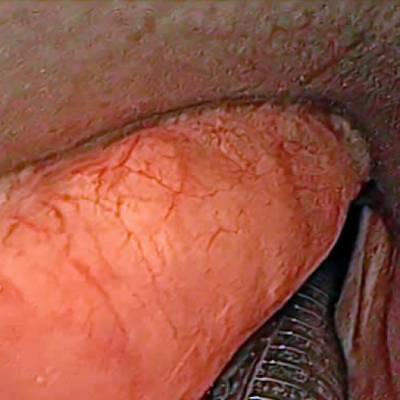

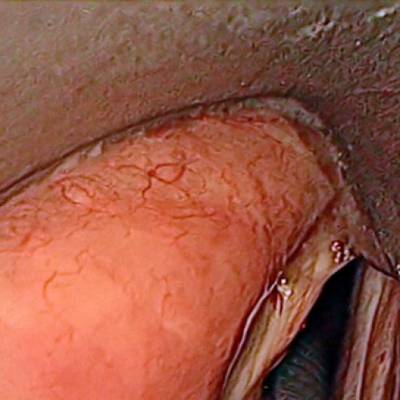

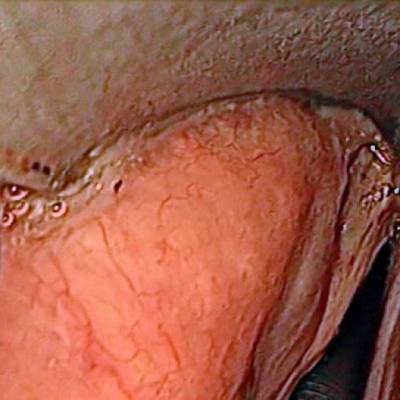

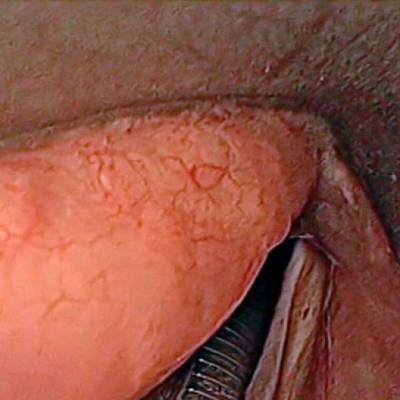

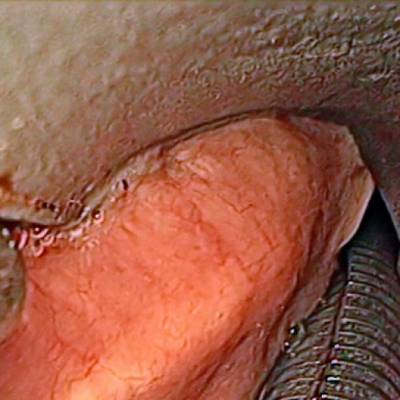

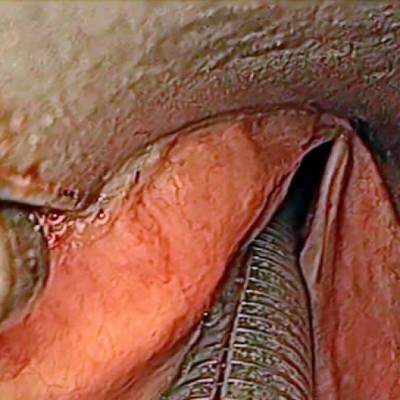

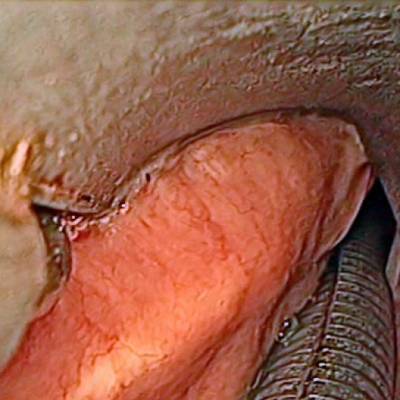

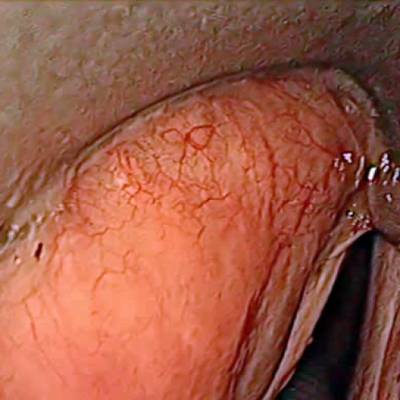

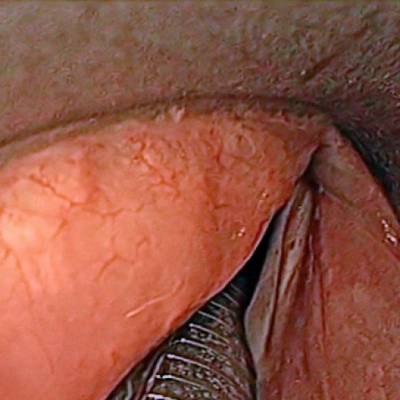

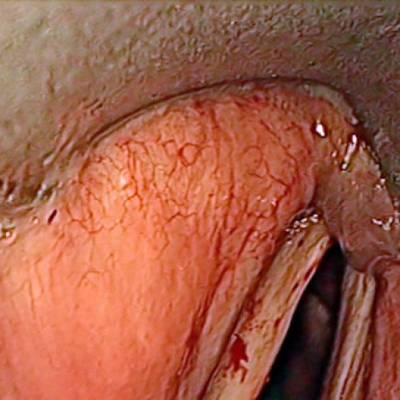

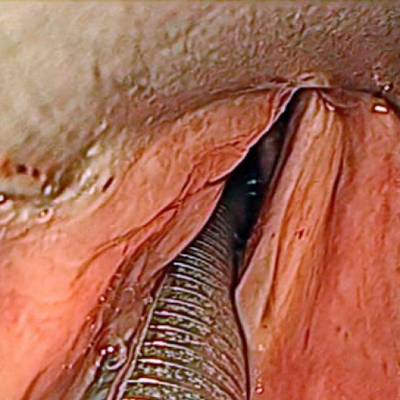

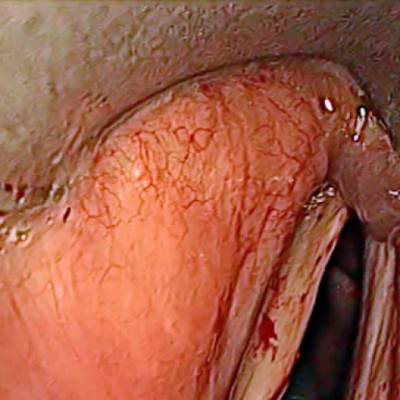

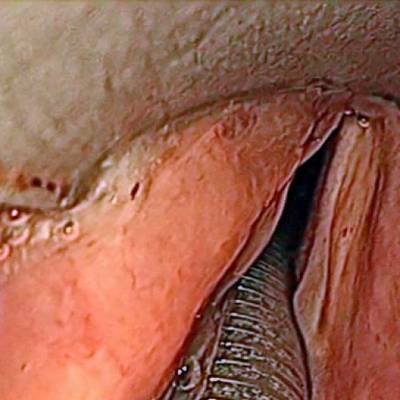

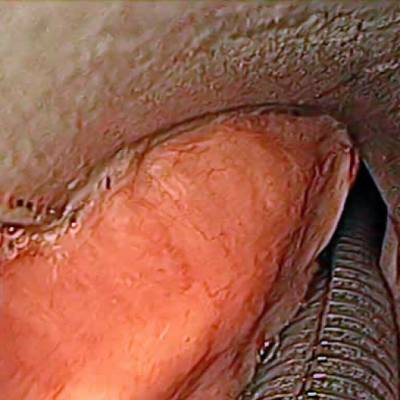

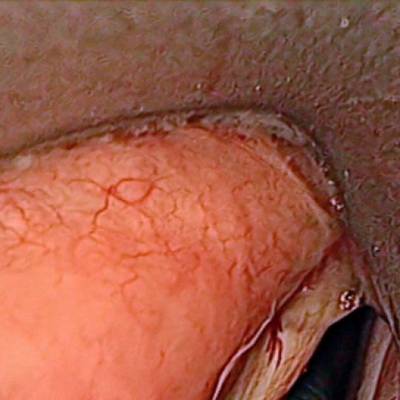

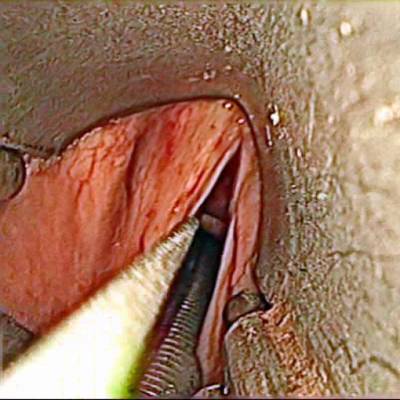

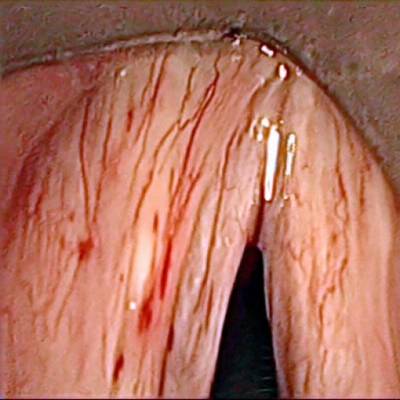

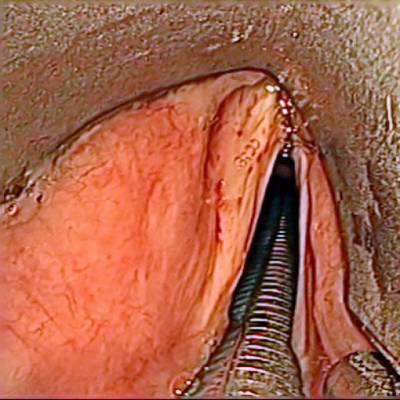

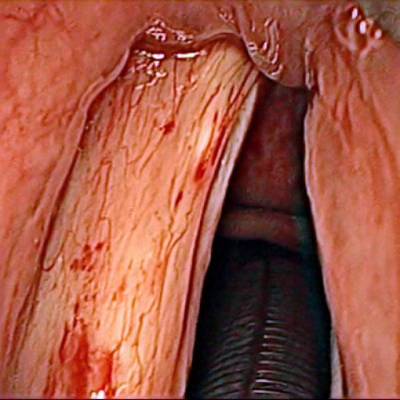
Class balance #
There are 7 annotation classes in the dataset. Find the general statistics and balances for every class in the table below. Click any row to preview images that have labels of the selected class. Sort by column to find the most rare or prevalent classes.
Class ㅤ | Images ㅤ | Objects ㅤ | Count on image average | Area on image average |
|---|---|---|---|---|
vocal folds➔ mask | 533 | 533 | 1 | 35.57% |
void➔ mask | 531 | 531 | 1 | 3.39% |
glottal space➔ mask | 529 | 529 | 1 | 8.15% |
other tissue➔ mask | 482 | 482 | 1 | 30.82% |
intubation➔ mask | 433 | 433 | 1 | 6.64% |
surgical tool➔ mask | 373 | 373 | 1 | 28.6% |
pathology➔ mask | 58 | 58 | 1 | 2.21% |
Co-occurrence matrix #
Co-occurrence matrix is an extremely valuable tool that shows you the images for every pair of classes: how many images have objects of both classes at the same time. If you click any cell, you will see those images. We added the tooltip with an explanation for every cell for your convenience, just hover the mouse over a cell to preview the description.
Images #
Explore every single image in the dataset with respect to the number of annotations of each class it has. Click a row to preview selected image. Sort by any column to find anomalies and edge cases. Use horizontal scroll if the table has many columns for a large number of classes in the dataset.
Object distribution #
Interactive heatmap chart for every class with object distribution shows how many images are in the dataset with a certain number of objects of a specific class. Users can click cell and see the list of all corresponding images.
Class sizes #
The table below gives various size properties of objects for every class. Click a row to see the image with annotations of the selected class. Sort columns to find classes with the smallest or largest objects or understand the size differences between classes.
Class | Object count | Avg area | Max area | Min area | Min height | Min height | Max height | Max height | Avg height | Avg height | Min width | Min width | Max width | Max width |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
vocal folds mask | 533 | 35.57% | 95.9% | 2.43% | 181px | 35.35% | 512px | 100% | 426px | 83.29% | 60px | 11.72% | 512px | 100% |
void mask | 531 | 3.39% | 33.42% | 0% | 1px | 0.2% | 512px | 100% | 376px | 73.52% | 1px | 0.2% | 512px | 100% |
glottal space mask | 529 | 8.15% | 92.7% | 0.05% | 4px | 0.78% | 512px | 100% | 245px | 47.9% | 13px | 2.54% | 512px | 100% |
other tissue mask | 482 | 30.82% | 67.41% | 0.26% | 38px | 7.42% | 512px | 100% | 420px | 82.08% | 28px | 5.47% | 512px | 100% |
intubation mask | 433 | 6.64% | 33.31% | 0.09% | 6px | 1.17% | 512px | 100% | 175px | 34.13% | 27px | 5.27% | 512px | 100% |
surgical tool mask | 373 | 28.6% | 88.48% | 0.1% | 19px | 3.71% | 512px | 100% | 383px | 74.83% | 23px | 4.49% | 512px | 100% |
pathology mask | 58 | 2.22% | 11.09% | 0.08% | 19px | 3.71% | 270px | 52.73% | 99px | 19.24% | 9px | 1.76% | 168px | 32.81% |
Spatial Heatmap #
The heatmaps below give the spatial distributions of all objects for every class. These visualizations provide insights into the most probable and rare object locations on the image. It helps analyze objects' placements in a dataset.
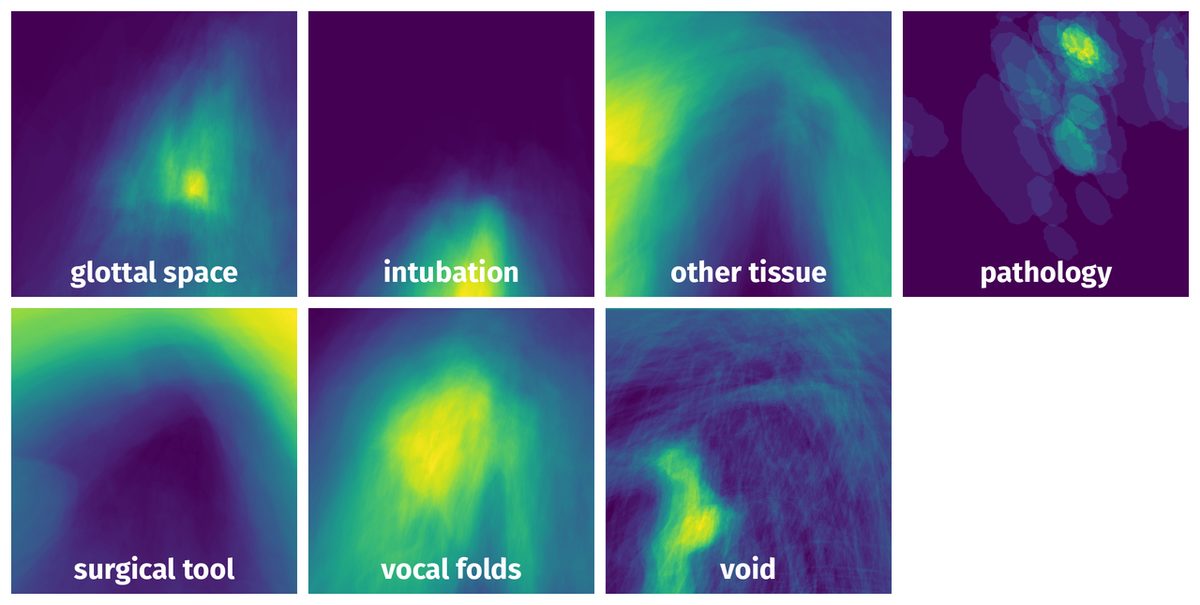
Objects #
Table contains all 2939 objects. Click a row to preview an image with annotations, and use search or pagination to navigate. Sort columns to find outliers in the dataset.
Object ID ㅤ | Class ㅤ | Image name click row to open | Image size height x width | Height ㅤ | Height ㅤ | Width ㅤ | Width ㅤ | Area ㅤ |
|---|---|---|---|---|---|---|---|---|
1➔ | void mask | seq6_0183_l.png | 512 x 512 | 512px | 100% | 512px | 100% | 13.06% |
2➔ | vocal folds mask | seq6_0183_l.png | 512 x 512 | 464px | 90.62% | 458px | 89.45% | 43.2% |
3➔ | other tissue mask | seq6_0183_l.png | 512 x 512 | 512px | 100% | 512px | 100% | 25.58% |
4➔ | glottal space mask | seq6_0183_l.png | 512 x 512 | 291px | 56.84% | 152px | 29.69% | 10.4% |
5➔ | intubation mask | seq6_0183_l.png | 512 x 512 | 154px | 30.08% | 166px | 32.42% | 7.76% |
6➔ | void mask | seq8_1085.png | 512 x 512 | 399px | 77.93% | 354px | 69.14% | 0.01% |
7➔ | vocal folds mask | seq8_1085.png | 512 x 512 | 471px | 91.99% | 402px | 78.52% | 39.5% |
8➔ | other tissue mask | seq8_1085.png | 512 x 512 | 512px | 100% | 512px | 100% | 46.04% |
9➔ | glottal space mask | seq8_1085.png | 512 x 512 | 74px | 14.45% | 72px | 14.06% | 1.05% |
10➔ | surgical tool mask | seq8_1085.png | 512 x 512 | 248px | 48.44% | 122px | 23.83% | 7.89% |
License #
Citation #
If you make use of the Laryngeal Endoscopic data, please cite the following reference:
@dataset{Laryngeal Endoscopic,
author={Max-Heinrich Laves and Jens Bicker and Lüder A. Kahrs and Tobias Ortmaier},
title={A Dataset of Laryngeal Endoscopic Images for Semantic Segmentation},
year={2020},
url={https://github.com/imesluh/vocalfolds}
}
If you are happy with Dataset Ninja and use provided visualizations and tools in your work, please cite us:
@misc{ visualization-tools-for-vocalfolds-dataset,
title = { Visualization Tools for Laryngeal Endoscopic Dataset },
type = { Computer Vision Tools },
author = { Dataset Ninja },
howpublished = { \url{ https://datasetninja.com/vocalfolds } },
url = { https://datasetninja.com/vocalfolds },
journal = { Dataset Ninja },
publisher = { Dataset Ninja },
year = { 2026 },
month = { feb },
note = { visited on 2026-02-24 },
}Download #
Dataset Laryngeal Endoscopic can be downloaded in Supervisely format:
As an alternative, it can be downloaded with dataset-tools package:
pip install --upgrade dataset-tools
… using following python code:
import dataset_tools as dtools
dtools.download(dataset='Laryngeal Endoscopic', dst_dir='~/dataset-ninja/')
Make sure not to overlook the python code example available on the Supervisely Developer Portal. It will give you a clear idea of how to effortlessly work with the downloaded dataset.
The data in original format can be downloaded here.
Disclaimer #
Our gal from the legal dep told us we need to post this:
Dataset Ninja provides visualizations and statistics for some datasets that can be found online and can be downloaded by general audience. Dataset Ninja is not a dataset hosting platform and can only be used for informational purposes. The platform does not claim any rights for the original content, including images, videos, annotations and descriptions. Joint publishing is prohibited.
You take full responsibility when you use datasets presented at Dataset Ninja, as well as other information, including visualizations and statistics we provide. You are in charge of compliance with any dataset license and all other permissions. You are required to navigate datasets homepage and make sure that you can use it. In case of any questions, get in touch with us at hello@datasetninja.com.


