Introduction #
The ISIC 2017: Part 3 - Disease Classification Task dataset is tailored for a classification task in the domain of dermatology. Comprising a total of 2750 images, the dataset presents two distinct binary classification tasks. The first task involves discriminating between nevus_or_seborrheic_keratosis, encompassing 2229 images, and melanoma, consisting of 521 images. The second task requires classification between melanoma_or_nevus, with 2364 images, and seborrheic_keratosis, containing 386 images. Participants in this task are tasked with independently addressing these two binary image classification challenges, each centered around three distinctive diagnoses of skin lesions: melanoma, nevus, and seborrheic keratosis. The dataset serves as a valuable resource for advancing the development and evaluation of algorithms dedicated to dermatological disease classification.
About ISIC 2017 Challenge
The International Skin Imaging Collaboration (ISIC) has begun to aggregate a large-scale publicly accessible dataset of dermoscopy images. Currently, the dataset houses more than 20,000 images from leading clinical centers internationally, acquired from a variety of devices used at each center. The ISIC dataset was the foundation for the first public benchmark challenge on dermoscopic image analysis in 2016.
The goal of the challenge is to help participants develop image analysis tools to enable the automated diagnosis of melanoma from dermoscopic images. Image analysis of skin lesions is composed of 3 parts:
- Part 1: Lesion Segmentation (Available on DatasetNinja)
- Part 2: Detection and Localization of Visual Dermoscopic Features/Patterns
- Part 3: Disease Classification (current)
For each, data consisted of images and corresponding ground truth annotations, split into training (n=2000), validation (n=150), and holdout test (n=600) datasets. Predictions could be submitted on validation and test datasets. The validation submissions provided instantaneous feedback in the form of performance evaluations, as well as ranking in comparison to other participants. Test submissions only provided feedback after the submission deadline.
About Disease Classification Task
In this task, participants are asked to complete two independent binary image classification tasks that involve three unique diagnoses of skin lesions (melanoma, nevus, and seborrheic keratosis). In the first binary classification task, participants are asked to distinguish between melanoma and nevus and seborrheic keratosis. In the second binary classification task, participants are asked to distinguish between seborrheic keratosis and nevus and melanoma.
- Melanoma – malignant skin tumor, derived from melanocytes (melanocytic)
- Nevus – benign skin tumor, derived from melanocytes (melanocytic)
- Seborrheic keratosis – benign skin tumor, derived from keratinocytes (non-melanocytic)
Example images from “Part 3: Disease Classification.” Ground truth labels written above.
Summary #
ISIC 2017: Part 3 - Disease Classification Task is a dataset for a classification task. It is used in the medical industry.
The dataset consists of 2750 images with 0 labeled objects. There are 3 splits in the dataset: train (2000 images), test (600 images), and valid (150 images). Alternatively, the dataset could be split into 2 groups for second classification task: melanoma_or_nevus (2364 images) and seborrheic_keratosis (386 images), or into 2 groups for first binary classification task: nevus_or_seborrheic_keratosis (2229 images) and melanoma (521 images). Also, the dataset contains age_approximate and sex tags. The dataset was released in 2017 by the IBM T. J. Watson Research Center, Emory University, University of Central Arkansas, Kitware, Memorial Sloan-Kettering Cancer Center, Missouri University of Science and Technology, USA, and Medical University of Vienna, Austria.
Here are the visualized examples for the classes:
Explore #
ISIC 2017: Part 3 - Disease Classification Task dataset has 2750 images. Click on one of the examples below or open "Explore" tool anytime you need to view dataset images with annotations. This tool has extended visualization capabilities like zoom, translation, objects table, custom filters and more. Hover the mouse over the images to hide or show annotations.

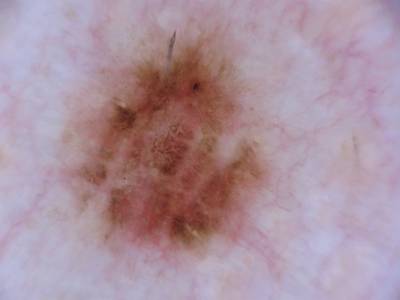

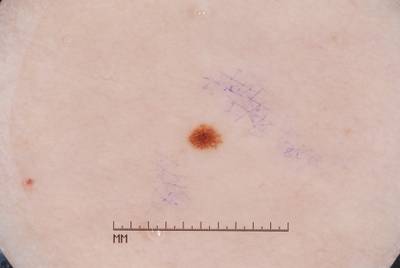

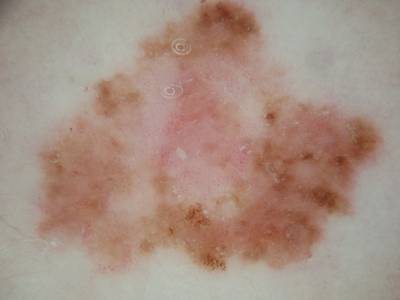

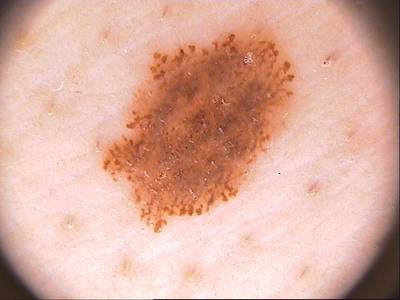

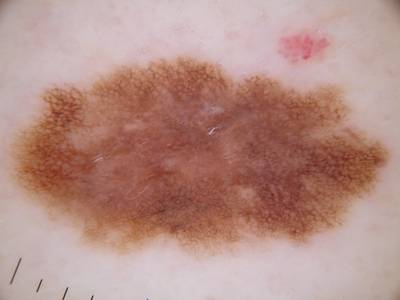





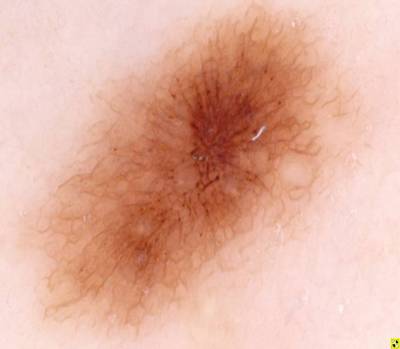

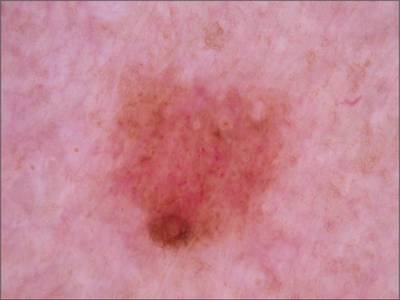

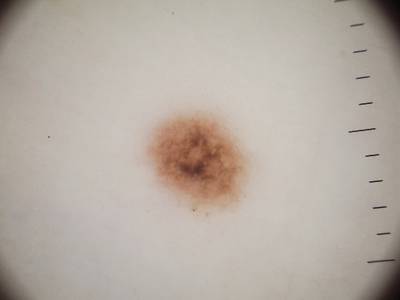



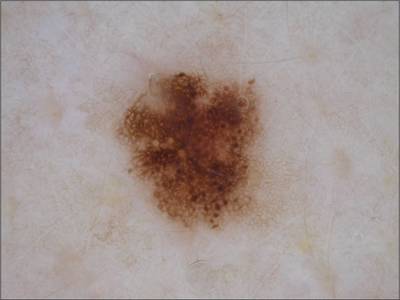



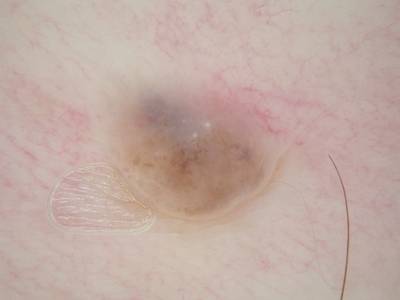

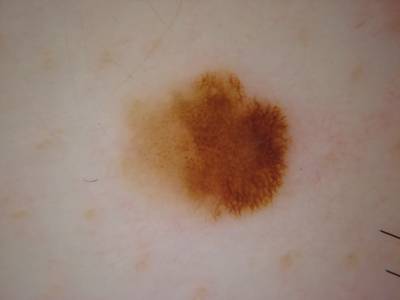

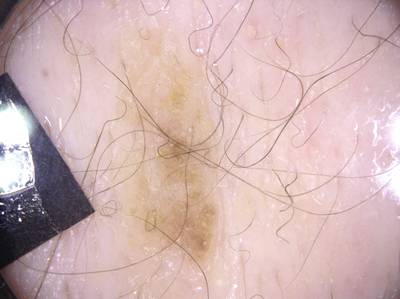

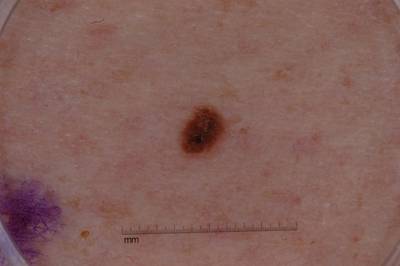

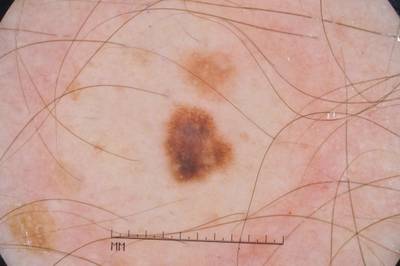

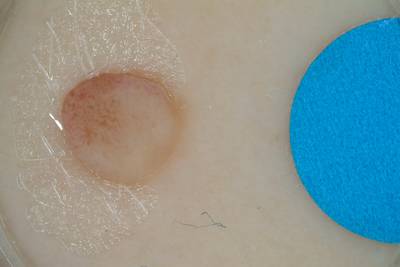

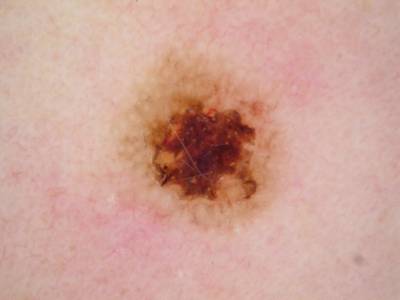



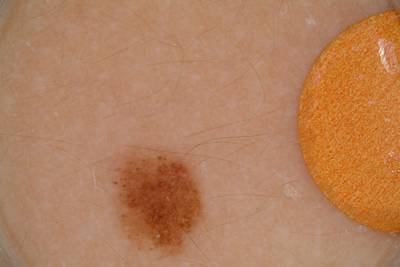

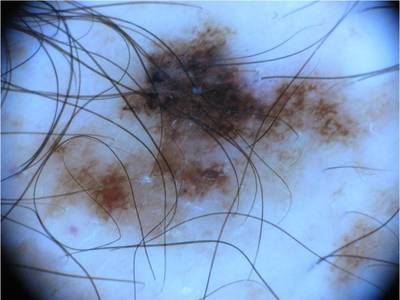

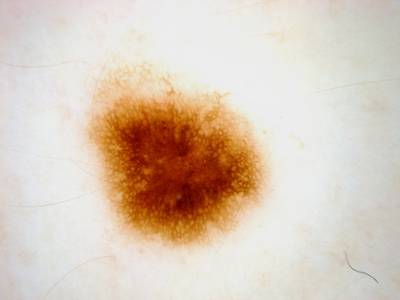



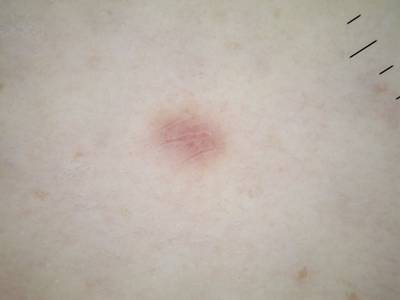

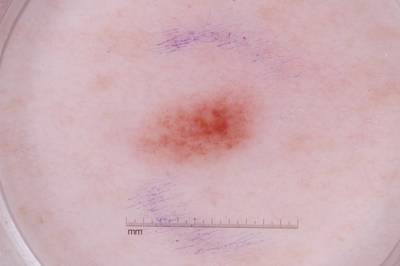

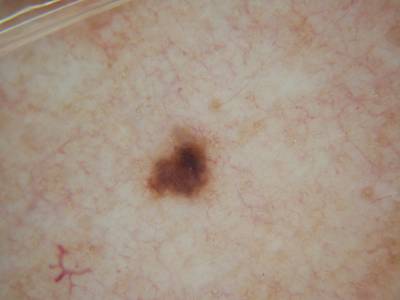

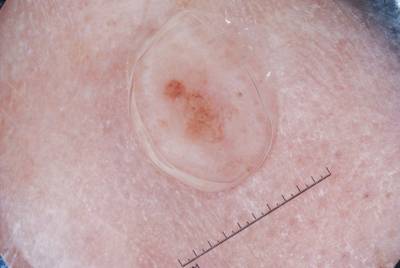

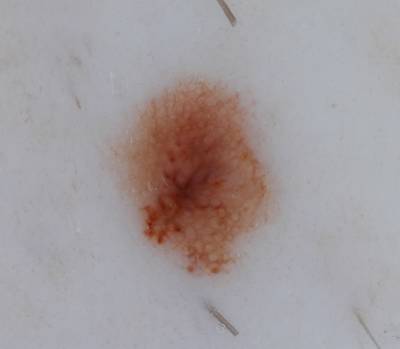
License #
Citation #
If you make use of the ISIC 2017: Part 3 - Disease Classification Task data, please cite the following reference:
@misc{codella2018skin,
title={Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC)},
author={Noel C. F. Codella and David Gutman and M. Emre Celebi and Brian Helba and Michael A. Marchetti and Stephen W. Dusza and Aadi Kalloo and Konstantinos Liopyris and Nabin Mishra and Harald Kittler and Allan Halpern},
year={2018},
eprint={1710.05006},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
If you are happy with Dataset Ninja and use provided visualizations and tools in your work, please cite us:
@misc{ visualization-tools-for-isic-2017-part-dataset,
title = { Visualization Tools for ISIC 2017: Part 3 - Disease Classification Task Dataset },
type = { Computer Vision Tools },
author = { Dataset Ninja },
howpublished = { \url{ https://datasetninja.com/isic-2017-part-3 } },
url = { https://datasetninja.com/isic-2017-part-3 },
journal = { Dataset Ninja },
publisher = { Dataset Ninja },
year = { 2026 },
month = { feb },
note = { visited on 2026-02-28 },
}Download #
Dataset ISIC 2017: Part 3 - Disease Classification Task can be downloaded in Supervisely format:
As an alternative, it can be downloaded with dataset-tools package:
pip install --upgrade dataset-tools
… using following python code:
import dataset_tools as dtools
dtools.download(dataset='ISIC 2017: Part 3 - Disease Classification Task', dst_dir='~/dataset-ninja/')
Make sure not to overlook the python code example available on the Supervisely Developer Portal. It will give you a clear idea of how to effortlessly work with the downloaded dataset.
The data in original format can be downloaded here.
Disclaimer #
Our gal from the legal dep told us we need to post this:
Dataset Ninja provides visualizations and statistics for some datasets that can be found online and can be downloaded by general audience. Dataset Ninja is not a dataset hosting platform and can only be used for informational purposes. The platform does not claim any rights for the original content, including images, videos, annotations and descriptions. Joint publishing is prohibited.
You take full responsibility when you use datasets presented at Dataset Ninja, as well as other information, including visualizations and statistics we provide. You are in charge of compliance with any dataset license and all other permissions. You are required to navigate datasets homepage and make sure that you can use it. In case of any questions, get in touch with us at hello@datasetninja.com.


